Results and Discussion - Identification of Drug Targets from Bacterial Genomoe
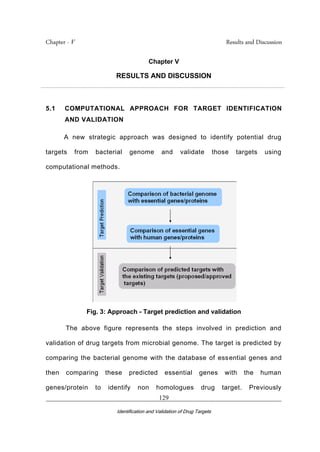
- 1. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 129 Chapter V RESULTS AND DISCUSSION . . 5.1 COMPUTATIONAL APPROACH FOR TARGET IDENTIFICATION AND VALIDATION A new strategic approach was designed to identify potential drug targets from bacterial genome and validate those targets using computational methods. Fig. 3: Approach - Target prediction and validation The above figure represents the steps involved in prediction and validation of drug targets from microbial genome. The target is predicted by comparing the bacterial genome with the database of essential genes and then comparing these predicted essential genes with the human genes/protein to identify non homologues drug target. Previously
- 2. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 130 subtractive genomics approach was used (Sakharkar et al., 2004; Anirban Dutta et al., 2006) to identify potential drug targets in Pseudomonas aeruginosa and Helicobacter pylori. In our approach the target identification and validation process is automated so that the user can submit the input (genome of a pathogenic microbe) and get the output as target sequences. The target sequences were analyzed for its functional role using sequence analysis tools (BLAST and Pfam). The validation of these drug targets were done by comparing these obtained against the approved and proposed genes/proteins from the Drugbank database. Target identification involves two steps as shown in the above Fig. 3. The essential genes in the microbes are identified by comparing them with the sequences of Database of Essential Genes. The genes which are homologous with the DEG are designated as essential genes. The approach involves comparing each gene from the genome and comparing them with the DEG database. The genes are compared based on the specified cut off valve and are stored in a text file. The text file would contain the gene sequences in fasta format. These matching genes will become the input for the next step.
- 3. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 131 Fig. 8: Screenshot of the web based tool The input genome sequence in text file format is uploaded in the region marked as ‘input reference files’. The database or set of sequences to be compared can be uploaded in the next region marked as ‘file to compare’. Once you have uploaded both these sequences, on clicking the submit button the tool compares each sequence from the input file sequence and compares with all the sequences in the ‘file to compare’ sequences. By default it compares these two set of sequences with the e-value of 1e–3 . These sequences can also be compared based on modifying the e-value cut offs.
- 4. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 132 Step-1: Comparison with Database of essential genes In the first step of comparison, the input genome sequences of a bacterial organism are compared with the database of essential genes. The sequence which matches with the DEG is written separately in a file. These genes are designated as essential genes to the bacterial species. This represents a pool of drug targets. Since drug discovery industry focuses on specific drug targets, these targets have to be drilled down to specific gene or protein target. This is achieved in the further steps in the algorithm. Step-2: Comparison with Human Homologue This step represents excluding human homologues. The target should not be homologous with humans and hence this step involves comparison of the essential genes predicted from the previous step with the human genes or proteins. The sensitivity and allergic reactions to the drug arises as a result of drug interfering with the host metabolic process apart from the target organism. If there is high level of stringency implemented in this step it can avoid lot of pit falls which may arise in the clinical trials. Most of the drug which has a reasonable biochemical effect often fails in the clinical testing as they interfere with the host mechanism. This is a very crucial step in the process of drug design and discovery. Now, the tool has to be run the second time to compare the input files (predicted bacterial essential genes) with the human genes sequences. To compare with the human genes for related sequences, they were downloaded from the NCBI ftp site.
- 5. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 133 The essential genes identified in step 1 are compared with the human genes. The genes which were homologous with the human genes are excluded in this step. These genes are designated as target genes. These genes were stored in a separate text file in fasta format. Fig. 8: Screenshot of the web based tool Step-3: Comparison with Approved /Predicted Targets The final step includes comparison of the target genes with the approved targets or already predicted targets to validate the findings. The predicted targets were validated by comparing them against the approved and proposed targets from DrugBank. DrugBank has more than 2500 non- redundant drug targets. The validation results reveal that most of the predicted targets using our approach fetched new targets when compared with the existing target database.
- 6. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 134 5.2 APPLICATION DEVELOPMENT Based on the designed approach a web based application was developed using Java. The application initially takes the input genome data and the essential genes in text file format or .ffn file format. Once it compares, the related sequences are retrieved in a separate text file in a specific location. These essential genes are then compared with the human genes. The comparison was carried out using BLAST program (BLASTall exe). BLAST executables were downloaded from NCBI site (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/) and it was customized to compare the input genome data with the essential genes and thereafter with the human genes to exclude the homologues. The web-based application was developed using JSP, Servlets and applying Struts framework. Using the developed application, the potential targets were identified for 80 pathogenic organisms and they were validated (Table-1).
- 7. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 135 5.2.1 Data Analysis for Target prediction and Validation Table-1 List of pathogenic organisms and predicted drug targets S.No. List of Pathogens Total number of genes in the genome Number of Potential TargetsProteins/Coding genes Proteins from plasmids 1 Acinetobacter baumannii AB0057 3790 11 91 2 Bacillus anthracis 5311 61 3 Bacillus subtilis 4177 162 4 Bacillus_cereus_ATCC_10987 5903 241 114 5 Bacteroides_fragilis_YCH46 4578 47 67 6 Bacteroides_fragilis_NCTC_9434 4184 47 86 7 Bartonella henselae 1488 67 8 Bordetella parapertussis 4185 95 9 Bordetella bronchiseptica 4994 88 10 Bordetella pertussis 3436 88 11 Burkholderia_mallei_ATCC_23344 5024 97 12 Brucella abortus 3000 80
- 8. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 136 S.No. List of Pathogens Total number of genes in the genome Number of Potential TargetsProteins/Coding genes Proteins from plasmids 13 Brucella suis 1330 3272 79 14 Chlamydia trachomatis 880 44 15 Chlamydophila_pneumoniae_AR39 1112 43 16 Clostridium botulinum 3548 90 17 Clostridium_difficile_630 3742 11 75 18 Clostridium perfringens 2558 20 76 19 Clostridium_perfringens_ATCC_13124 2876 78 20 Clostridium_tetani_E88 2373 59 71 21 Coxiella_burnetii_RSA_331 1930 45 79 22 Corynebacterium diphtheriae 2272 42 23 Campylobacter_fetus_82-40 1719 92 24 Campylobacter jejuni 1838 93 25 Ehrlichia_chaffeensis_Arkansas 1105 44 26 Escherichia_coli_K_12_substr__MG1655 4149 164 27 Escherichia_coli_UTI89 5021 145 184 28 Francisella tularensis 1754 76 29 Haemophilus influenzae 1792 452
- 9. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 137 S.No. List of Pathogens Total number of genes in the genome Number of Potential TargetsProteins/Coding genes Proteins from plasmids 30 Helicobacter pylori 1489 242 31 Klebsiella pneumoniae 5425 343 158 32 Listeria monocytogenes 2846 86 33 Listeria_monocytogenes_Clip81459 2766 86 34 Listeria_monocytogenes_HCC23 2974 85 35 Leptospira interrogans 4724 81 36 Leptospira_interrogans_serovar_Copenhageni 3658 81 37 Leptospira_biflexa_serovar_Patoc__Patoc_1__Ames 3667 59 79 38 Mycobacterium leprae 1605 52 39 Mycobacterium tuberculosis 3989 44 40 Mycobacterium_tuberculosis_F11 3941 53 41 Mycobacterium_tuberculosis_H37Ra 4034 53 42 Mycoplasma pneumoniae 689 151 43 Mycoplasma genitalium 475 220 44 Neisseria gonorrhoeae 2002 81 45 Neisseria meningitidis 1917 84 46 Pasteurella multocida 2015 170
- 10. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 138 S.No. List of Pathogens Total number of genes in the genome Number of Potential TargetsProteins/Coding genes Proteins from plasmids 47 Proteus mirabilis 3607 55 123 48 Propionibacterium_acnes_KPA171202 2297 61 49 Psendomonas aeruginosa 5566 109 50 Rickettsia_rickettsii_Iowa 1384 62 51 Rickettsia_akari_Hartford 1259 57 52 Salmonella_enterica_Paratypi_ATCC_9150 4093 148 53 Salmonella_enterica_serovar_Typhi_Ty2 4318 148 54 Serratia_proteamaculans_568 4891 51 148 55 Streptococcus_pyogenes_MGAS10270 1986 64 56 Salmonella typhimurium 4423 102 152 57 Staphylococcus_aureus_JH9 2697 29 117 58 Staphylococcus_epidermidis_ATCC_12228 2419 66 85 59 Shigella dyseneriae 4271 231 153 60 Stenotrophomonas_maltophilia_K279a 4386 92 61 Streptococcus pneumoniae 2202 72 62 Treponema pallidum 1028 33 63 Ureaplasma urealyticum 646 53
- 11. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 139 S.No. List of Pathogens Total number of genes in the genome Number of Potential TargetsProteins/Coding genes Proteins from plasmids 64 Vibrio cholerae 3693 121 65 Vibrio_parahaemolyticus 4832 133 66 Vibrio_vulnificus_CMCP6 4472 118 67 Wolinella_succinogenes 2042 116 68 Yersinia enterocolitica 3979 72 147 69 Yersinia pseudotuberculosis 4124 200 136 70 Yersinia pestis_KIM 4054 116 137 71 Clostridium_perfringens str 13 2660 63 76 72 Clostridium_acetobutylicum 3672 176 97 73 Desulfovibrio_vulgaris_DP4 2941 150 76 74 Microcystis_aeruginosa_NIES_843 6312 64 75 Pseudomonas aeruginosa PA7 6286 123 76 Acidobacterium_capsulatum_ATCC_51196 3377 80 77 Chlamydia_trachomatis_L2b_UCH_1_proctitis 874 46 78 Staphylococcus_aureus_COL 2612 3 116 79 Staphylococcus_aureus_Mu50 2697 34 110 80 Staphylococcus aureus subsp. aureus N315 2588 31 114
- 12. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 140 The table shows the number of targets predicted for selected pathogenic organisms. A total of 8171 drug targets were predicted from these 80 organisms. The minimal number of targets were found in Treponema pallidum (33 targets) and the maximum target were found in Haemophilus influenza (452 targets). The predicted targets were organized in a web based database. 5.2.2 Case scenario – Mycobacterium tuberculosis Tuberculosis has re-emerged as a global health concern due to declining efficiency of current therapeutic agents and development of multi drug resistant strains of Mycobacterium tuberculosis. The currently used drug combination is no longer considered an eternal solution for treating the disease. These drugs were originally discovered and formulated in 1940’s and it’s still in the clinician’s prescription. Due to advancements in genome sequence technologies, the current research has resulted in few clinical trials. In 1938 the complete genome sequence of M.tuberculosis was completed. Since then numerous initiatives are carried out using the genome data to identify TB drug targets. Growing concern and potential solutions Nowadays, about 70% of the bacteria that cause infections in hospitals are resistant to at least one of the antibiotic agents most commonly used for treatment. Some organisms are resistant to all approved antibiotics and can only are treated with experimental and potentially toxic drugs.
- 13. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 141 Factors causing resistance Incorrect use of antibiotics Patient related factors Prescriber’s prescription Use of monotherapy Commercial promotion Over the counter sale of antibiotics Under use of microbiological testing and globalization Incorrect use of antibiotics such as too short a time, at too low a dose, at inadequate potency or for the wrong diagnosis always enhances the likelihood of bacterial resistance to these drugs. Due to the selection pressure caused by antibiotic use, a large pool of resistant genes has been created and this antibiotic resistance places an increased burden on society in terms of high morbidity, mortality and cost. As a whole antibiotic resistance increases the healthcare cost, increasing the severity of disease and death rates of few infections. CDC has estimated that some 150 million prescriptions every year are unnecessary. The analysis of the Mycobacterium tuberculosis genome data using our application showed 53 potential targets. These targets were analysed for their conservity among other organisms using blast searchers and the results are tabulated.
- 14. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 142 Table-2 Validated Drug Targets from Mycobacterium tuberculosis S. № Target protein Conservity 1. Cell division protein rodA Conserved only among the Mycobacterial organisms. 2. Cell division protein FtsA Conserved only among the Mycobacterial organisms. 3. Replicative DNA helicase Conserved among the Mycobacterial and few other organisms. 4. Dihydroxy-acid dehydratase Conserved only among the Mycobacterial organisms.. 5. Fructose-bisphosphate aldolase fba Conserved among the Mycobacterial and few other organisms. 6. Transcription antitermination protein nusG Conserved among the Mycobacterial and few other organisms. 7. 50S ribosomal protein L1 rplA Conserved among the Mycobacterial and few other organisms. 8. 30S ribosomal protein S19 rpsS and 50S ribosomal protein L22 rplV Conserved among the Mycobacterial and few other organisms. 9. 50S ribosomal protein L22 rplV and 30S ribosomal protein S3 rpsC Conserved among the Mycobacterial and few other organisms. 10. 50S ribosomal protein L24 rplX and 50S ribosomal protein L5 rplE Conserved among the Mycobacterial and few other organisms. 11. 30S ribosomal protein S8 rpsH Conserved among the Mycobacterial organisms and Streptomyces griseus subsp. griseus NBRC 13350 12. 30S ribosomal protein S5 rpsE Conserved only among the Mycobacterial organisms.
- 15. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 143 S. № Target protein Conservity 13. Preprotein translocase subunit secY Conserved only among the Mycobacterial organisms. 14. Acetyl-CoA carboxylase carboxyl transferase beta subunit accD3 Conserved only among the Mycobacterial organisms. 15. lytB-related protein lytB2 Conserved only among the Mycobacterial organisms. 16. Conserved hypothetical protein excinuclease ABC subunit C uvrC Conserved only among the Mycobacterial organisms. 17. Conserved hypothetical protein Conserved only among the Mycobacterial organisms. 18. DNA polymerase subunit III alpha dnaE1 Conserved only among the Mycobacterial and few other organisms. 19. Drug efflux membrane protein Conserved only among the Mycobacterial and few other pathogenic organisms. 20. Initiation factor IF-3 infC Conserved only among the Mycobacterial organisms. 21. Phenylalanyl-tRNA synthetase subunit beta pheT and phenylalanyl-tRNA synthetase subunit alpha pheS Conserved only among the Mycobacterial organisms. 22. Cytotoxin/hemolysin and inorganic polyphosphate/ATP- NAD kinase- Conserved only among the Mycobacterial organisms. 23. ScpA/B family protein and initiation inhibitor protein Conserved only among the Mycobacterial organisms. 24. Preprotein translocase ATPase subunit secA2 Conserved only among the Mycobacterial and few other pathogenic organisms. This target sequence matches with the already approved target sequences from drug bank.
- 16. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 144 S. № Target protein Conservity 25. UDP-N-acetylmuramate-alanine ligase MurC Conserved only among the Mycobacterial organisms. 26. UDP-N-acetylglucosamine-N- acetylmuramyl- (pentapeptide)pyrophosphoryl- undecaprenol-N- acetylglucosamine transferase MurG Conserved only among the Mycobacterial organisms. 27. Cell division protein ftsW Conserved only among the Mycobacterial organisms. 28. UDP-N-acetylmuramoylalanine- D-glutamate ligase MurD Conserved only among the Mycobacterial and few other organisms. 29. Phospho-N-acetylmuramoyl- pentappeptidetransferase MurX Conserved only among the Mycobacterial organisms. 30. Phospho-N-acetylmuramoyl- pentapeptide-transferase and UDP-N-acetylmuramoylalanyl- D-glutamyl-2,6-diaminopimelate- D-alanyl-D-alanyl ligase Conserved only among the Mycobacterial organisms. 31. UDP-N-acetylmuramoylalanyl- D-glutamate-2,6-diaminopimelat E ligase MurE and UDP-N- acetylmuramoylalanyl-D- glutamyl-2, 6- diaminopimelate-D-alanyl-D- alanyl ligase MurF Conserved only among the Mycobacterial organisms. 32. Methylase MraW, conserved proline rich membrane protein and penicillin-binding membrane protein pbpB Conserved only among the Mycobacterial and few other organisms. 33. Nicotinate-nucleotide adenylyltransferase nadD Conserved only among the Mycobacterial organisms. 34. Ribonuclease E rne and C4- dicarboxylate-transport transmembrane protein dctA Conserved only among the Mycobacterial and few other organisms.
- 17. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 145 S. № Target protein Conservity 35. Glyoxalase II and histidyl-tRNA synthetase hiss Conserved only among the Mycobacterial and few other organisms. 36. N utilization substance protein A nusA Conserved only among the Mycobacterial and few other organisms. 37. 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase gcpE Conserved only among the Mycobacterial organisms. 38. Uridylate kinase pyrH Conserved only among the Mycobacterial organisms. 39. 50S ribosomal protein L19 rplS Conserved only among the Mycobacterial organisms and few pathogenic organisms. 40. tRNA (guanine-N(1))- methyltransferase trmD Conserved only among the Mycobacterial organisms. 41. Phosphopantetheine adenylyltransferase kdtB Conserved only among the Mycobacterial organisms. 42. ATP-dependent DNA helicase recG Conserved only among the Mycobacterial organisms. 43. ATP-dependent DNA helicase II uvrD2 Conserved only among the Mycobacterial organisms. 44. ATP-dependent DNA helicase II uvrD2 Conserved only among the Mycobacterial organisms. 45. Preprotein translocase subunit Conserved only among the Mycobacterial organisms and few pathogenic organisms. 46. Uracil phosphoribosyltransferase upp Conserved only among the Mycobacterial organisms. 47. Error-prone DNA polymerase Conserved only among the Mycobacterial and few other organisms. 48. 1-deoxy-D-xylulose-5-phosphate synthase lytB-related protein lytB1 Conserved among the Mycobacterial organisms and all major pathogenic organisms.
- 18. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 146 S. № Target protein Conservity 49. DNA-directed RNA polymerase subunit alpha rpoA Conserved only among the Mycobacterial organisms and few pathogenic organisms. 50. translation initiation factor IF-1 infA Conserved only among the Mycobacterial organisms. 51. alpha, alpha-trehalose- phosphate synthase otsA Conserved only among the Mycobacterial organisms and few pathogenic organisms. 52. aspartate-semialdehyde dehydrogenase asd Conserved only among the Mycobacterial organisms and few pathogenic organisms. 53. Bifunctional UDP- galactofuranosyl transferase glfT and UDP-galactopyranose mutase glf Conserved only among the Mycobacterial organisms and few pathogenic organisms. UDP- galactopyranose mutase glf matches with the already approved target sequences from drug bank. Most of the targets predicted from the organism were new compared to the approved targets from the Drug Bank. Of the 53 targets obtained from Mycobacterium tuberculosis only two targets (Preprotein translocase ATPase subunit secA2 and Bifunctional UDP-galactofuranosyl transferase glfT and UDP-galactopyranose mutase glf were matching with the drug bank. Sequencing of bacterial genomes has been progressing with breathtaking speed. Industrial research is now facing the challenge of translating this information efficiently into drug discovery. Complete genome sequences of bacterial organisms have revolutionized the search
- 19. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 147 for antibiotics. The search for new antibiotics can be assisted by computational methods such as homology-based analyses, structural genomics, motif analyses, protein-protein interactions, and experimental functional genomics (Loferer, 2000). The greatest success of computer-aided structure-based drug design to date is the HIV-1 protease inhibitors that have been approved by the United States Food and Drug Administration and reached the market (Wlodawer and Vondrasek., 1998). There have been many successful computer-assisted molecular design attempts to involve the use of QSAR to improve activity of lead compounds. An example of the success story is that of SAR work carried out on antibacterial agent, Norfloxacin (Koga et al., 1980) that showed 6-fluro derivative of norfloxacin being 500 fold more potent over nalidixic acid. Other examples of drugs that were developed using computer assisted drug design include Captopril (antihypertensive), Crixican (anti-HIV) (Greer et al., 1994), Teveten (antihypertensive) (Keenan, 1993), Aricept (for Alzheimers disease) (Kawakami et al., 1996), Trusopt (for Glaucoma) (Greer et al., 1994) and Zomig (for migraine) (Glen et al., 1995). Similarly applying CADD concepts for these new targets will results in development of novel therapeutics as well as to manage multi-drug resistance. The database developed using the targets will serve as a key resource to facilitate drug design and discovery.
- 20. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 148 The data analysis was performed for a selected list of 80 pathogenic microbes. The average time taken for screening 2000 gene sequences was found to be 60 minutes. Though the developed approach was used to analyze these 80 organisms, a special emphasis was given for the Mycobacterium tuberculosis as it is a highly drug resistant organism. A comprehensive data analysis was performed for Mycobacterium tuberculosis. The predicted targets were analyzed for its functional role using bioinformatics tools. The target sequences like gene name, protein product, function, EC. NO, pathway were retrieved from the sequence database and separately populated in a web based database developed using JSP. This web based database will be made available free for the educational research institutions to promote discovery and development of novel drugs. 5.3 DATABASE DEVELOPMENT Database of bacterial drug targets The predicted targets from the selected pathogenic organism’s gene name, protein product, Enzyme Commission Number, function, functional information were collected and populated in a web based database to act as a reservoir for drug discovery.
- 21. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 149 Database Development Details Fig. 9: Screenshot of the database input screen Figure-9 shows the input screen for the database. The input data can be provided manually or as a single upload in a spreadsheet. The implementation of AJAX concepts for the search process renders effective querying methods and retrieves the results faster.
- 22. Chapter - V Results and Discussion _________________________________________________________________________ Identification and Validation of Drug Targets 150 Figure-10: Screenshot of the database screen Figures 9 and 10 shows the database input screen and the data updated in the database. The database also has option to upload the data directly from a Microsoft spreadsheet. The present research pursuit was initiated owing to the prevalence of multi-drug resistance and the pressing need for new drugs. Resistance is more likely when newly introduced antibiotics are chemically similar to ones already rendered ineffective. Therefore, new antimicrobial compounds should ideally have novel mechanisms of action. This demands design and development of compounds which is different in structure and mechanisms of action. Hence a new approach in drug design and discovery would eventually lead to novel class of drugs. _____
