Network cheminformatics: gap filling and identifying new reactions in metabolic networks based on metabolite similarity
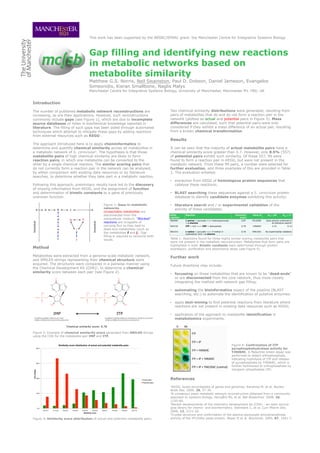
- 1. This work has been supported by the BBSRC/EPSRC grant: the Manchester Centre for Integrative Systems Biology Gap filling and identifying new reactions in metabolic networks based on metabolite similarity Matthew G.S. Norris, Neil Swainston, Paul D. Dobson, Daniel Jameson, Evangelos Simeonidis, Kieran Smallbone, Naglis Malys Manchester Centre for Integrative Systems Biology, University of Manchester, Manchester M1 7ND, UK Introduction The number of published metabolic network reconstructions are Two chemical similarity distributions were generated, resulting from increasing, as are their applications. However, such reconstructions pairs of metabolites that do and do not form a reaction pair in the commonly include gaps (see Figure 1), which are due to incomplete network (plotted as actual and potential pairs in Figure 3). Mass source databases or holes in biochemical knowledge reported in differences are calculated, such that potential pairs were only literature. The filling of such gaps has been aided through automated considered if they exhibit a mass difference of an actual pair, resulting techniques which attempt to mitigate these gaps by adding reactions from a known chemical transformation. from external resources such as KEGG1. Results The approach introduced here is to apply cheminformatics to determine and quantify chemical similarity across all metabolites in It can be seen that the majority of actual metabolite pairs have a a metabolic network of S. cerevisiae2. The hypothesis is that those chemical similarity score greater than 0.7. However, only 8.5% (557) metabolite pairs of high chemical similarity are likely to form of potential pairs exhibit such similarity. Of these 557, 99 were reaction pairs, in which one metabolite can be converted to the found to form a reaction pair in KEGG, but were not present in the other by a single chemical reaction. The similar scoring pairs that metabolic network. From these 99 pairs, a number were selected for do not currently form a reaction pair in the network can be analysed, further evaluation, and three examples of this are provided in Table by either comparison with existing data resources or by literature 1. The evaluation entailed: searches, to determine whether they take part in a metabolic reaction. • extraction from KEGG of homologous protein sequences that Following this approach, preliminary results have led to the discovery catalyse these reactions; of missing information from KEGG, and the assignment of function and determination of kinetic constants to a gene of previously • BLAST searching these sequences against a S. cerevisiae protein unknown function. database to identify candidate enzymes exhibiting this activity; Figure 1: Gaps in metabolic • literature search and / or experimental validation of the networks. activity of these candidates. Unreachable metabolites are disconnected from the KEGG Reaction Similarity Gene id KM / µM Kcat / s-1 reaction score extracellular medium. “Blocked” R00585 L-serine + pyruvate <=> hydroxypyruvate 0.87 YFL030W Gene activity confirmed in reactions are incapable of + L-alanine by literature search4 carrying flux as they lead to R00720 ITP + H2O <=> IMP + diphosphate 0.78 YJR069C 2.33 0.14 dead-end metabolites (such as R01215 L-valine + pyruvate <=> 3-methyl-2- 0.76 YER152C No experimental validation the metabolites f and j). Gap oxobutanoic acid + L-alanine filling is required to reconcile both issues. Table 1: Reactions found for three highly similar scoring metabolite pairs that were not present in the metabolic reconstruction. Metabolites that form pairs are highlighted in bold. Kinetic constants were determined through protein Method expression, purification and absorbance assay (see Figure 4). Metabolites were extracted from a genome-scale metabolic network, Further work and SMILES strings representing their chemical structure were acquired. The structures were compared in a pairwise manner using Future directions may include: the Chemical Development Kit (CDK)3, to determine a chemical similarity score between each pair (see Figure 2). • focussing on those metabolites that are known to be “dead-ends” or are disconnected from the core network, thus more-closely integrating the method with network gap filling; • automating the bioinformatics aspect of the pipeline (BLAST searching, etc.) to automate the identification of putative enzymes; • apply text-mining to find potential reactions from literature where reactions are not present in existing data resources such as KEGG; • application of the approach to metabolite identification in metabolomics experiments. Figure 2: Example of chemical similarity score generated from SMILES strings using the CDK for the metabolite pair IMP and ITP. Similarity score distribution of actual and potential metabolite pairs Figure 4: Confirmation of ITP 40.0 pyrophosphohydrolase activity for YJR069C. A Malachite Green assay was performed to detect orthophosphate, indicating hydrolysis of ITP and release 30.0 of pyrophosphate by YJR069C, which is further hydrolysed to orthophosphate by inorganic phosphatase (IP). ! Percentage 20.0 Actual pairs References Potential pairs 1KEGG: kyoto encyclopedia of genes and genomes. Kanehisa M, et al. Nucleic Acids Res. 2000, 28, 27-30. 10.0 2A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Herrgård MJ, et al. Nat Biotechnol. 2008, 26, 1155-60. 3Recent developments of the chemistry development kit (CDK) - an open-source 0.0 java library for chemo- and bioinformatics. Steinbeck C, et al. Curr Pharm Des. 0.0-0.1 0.1-0.2 0.2-0.3 0.3-0.4 0.4-0.5 0.5-0.6 Similarity score 0.6-0.7 0.7-0.8 0.8-0.9 0.9-1.0 2006, 12, 2111-20. 4Crystal structure and confirmation of the alanine:glyoxylate aminotransferase Figure 3: Similarity score distribution of actual and potential metabolite pairs. activity of the YFL030w yeast protein. Meyer P, et al. Biochimie. 2005, 87, 1041-7.
