Clam DEG
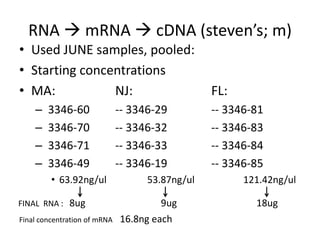
- 1. RNA mRNA cDNA (steven’s; m) • Used JUNE samples, pooled: • Starting concentrations • MA: NJ: FL: – 3346-60 -- 3346-29 -- 3346-81 – 3346-70 -- 3346-32 -- 3346-83 – 3346-71 -- 3346-33 -- 3346-84 – 3346-49 -- 3346-19 -- 3346-85 • 63.92ng/ul 53.87ng/ul 121.42ng/ul FINAL RNA : 8ug 9ug 18ug Final concentration of mRNA 16.8ng each
- 2. RNA cDNA (Jackie’s 1; c) • Used AUG samples, pooled • Starting concentrations: • MA: NJ: FL: – 3387-38 10ul -- 3387-1 10ul -- 3387-20 10ul – 3387-37 10ul -- 3387-25 10ul -- 3387-33 10ul – 3387-39 10ul -- 3387- 4 10ul -- 3387-28 10ul 85.47ng/ul 71.48ng/ul 51.80ng/ul LOWEST was FL: 7.5ul NJ: 5.4ul MA: 5.3ul for total amt of 388ng Still have 20ul from each
- 3. RNA cDNA (Jackie’s 2; c) • Used AUG samples, different amts for more concentrated pools • MA: NJ: FL: – 3387-37 15ul -- 3387-1 8ul -- 3386-33 15ul – 3387-39 15ul -- 3387-4 22ul -- 3386-28 15ul 85.47ng/ul 77.49ng/ul 99.25ng/ul LOWEST was NJ: 7.5ul MA: 6.79ul FL: 5.85ul for a total amt of 581ng Still have 20ul from each pool
- 4. PCRs with Steven’s samples • 6ul rxn with primers 7, 8, 59, 60 3.16.08 • 5ul rxn with primers 47-58 3.17.08 • 5ul rxn with primers 18, 20 3.18.08 • 3ul rxn with primers 1-6 3.?.08
- 5. PCRs with Jackie’s samples • LIBRARY 1 • 5ul rxn with primers 41-46 3.16.08 • 4ul rxn with primers 47-53 3.17.08 • LIBRARY 2 • 5ul rxn with primers 54-60 3.17.08 • 7ul rxn with primers 12,14,15,16,17,18,203.18.08
- 6. STEVEN’S SAMPLES (have m beside strain name) 52 53 54 55 56 57 Mm Nm Fm Mm Nm Fm Mm Nm Fm Mm Nm Fm Mm Nm Fm Mm Nm Fm 47 48 49 50 51 58 Mm Nm Fm Mm Nm Fm Mm Nm Fm Mm Nm Fm Mm Nm Fm Mm Nm Fm
- 7. STEVEN’S SAMPLES CONT 7 8 59 60 Fm Mm Nm Fm Mm Nm Fm Mm Nm Fm Mm Nm
- 8. STEVEN’S SAMPLES • Bands in red have been cut but not cloned • Bands in blue have been cut and cloned • CUT CLONED BAND FROZEN • -FLm 8a* -FLm 8a • -FLm 8b* -FLm 8b • -FLm 48 -FLm 48 • -FLm 49 -FLm 49 • -NJm 51* -NJm 51 • -NJm 52 -NJm52 • -FLm 52 -FLm 52 • -MAm 54 -MAm 54 • -FLm 55 -FLm 55 • -FLm 57 -FLm 57 • -FLm 58 -FLm 58
- 9. JACKIE’S SAMPLES (have c after strain name) 41 42 43 44 45 46 Mc Nc Fc Mc Nc Fc Mc Nc Fc Mc Nc Fc Mc Nc Fc Mc Nc Fc 47 48 49 50 51 52 53 Mc Nc Fc Mc Nc Fc Mc Nc Fc Mc Nc Fc Mc Nc Fc Mc Nc Fc Mc Nc Fc Plus NJc 12
- 10. JACKIE’S SAMPLES • Bands in red were cut but not cloned • Bands in blue were cut and cloned • CUT CLONED BAND FROZEN • -FLc 42a -FLc 42a • -FLc 42b -FLc 42b • -NJc 43 -NJc 43 • -NJc 46a* -NJc 42a • -NJc 46b* -NJc 42b • -NJc 48* -NJc 48 • -FLc 50* -FLc 50 • -NJc 51 -NJc 51 • -NJc 12 -NJc 12
- 11. COLONY PCR *denotes colonies selected for plasmid prep NJc 48 FLc 50 * * * NJc 46a 1 2 3 4 5 6 7 8 1 2 3 4 5 6 7 8 1 2 3 4 5 6 7 8 NJc 46b FLm 8a FLm 8b * * * * * * * 1 2 3 4 5 6 7 8 1 2 3 4 5 6 7 8 1 2 3 4 5 6 7 8 NJm 51 Clam prost. rec AB 21 * * * * * 1 2 3 4 5 6 7 8 1 2 3 4 5 6 7 8 1 2 3 4 5 6 7 8
- 12. Colonies taken for plasmid prep • NJc 48: 1, 2 • FLc 50: 4 • NJc 46a: 1, 2, 3, 4 *new* restreaked • NJc 46b: 1, 2, 3, 4 *new* restreaked • FLm 8a: 5, 8 • FLm 8b: 1, 2, 3, 4, 5 • NJm 51: 3, 4 • Pro recp: 6 • AB 21: 1, 7