Puton bosc2010 bio_python-modules-rna
•
1 recomendación•701 vistas
Denunciar
Compartir
Denunciar
Compartir
Descargar para leer sin conexión
Recomendados
Recomendados
http://www.baieuskarari.eus/es/comunidad-bai-euskarari/manual-de-practicas-ejemplares-en-euskara
http://www.baieuskarari.eus/fr/communaute-bai-euskarari/une-pratique-adequate-et-exemplaire-en-euskaraManual de prácticas ejemplares en euskara / Une pratique adéquate et exemplai...

Manual de prácticas ejemplares en euskara / Une pratique adéquate et exemplai...Bai Euskarari Ziurtagiriaren Elkartea
Más contenido relacionado
La actualidad más candente
La actualidad más candente (7)
Mohamed Ezzat El Zowalaty avian influenza PhD_thesis results

Mohamed Ezzat El Zowalaty avian influenza PhD_thesis results
Destacado
http://www.baieuskarari.eus/es/comunidad-bai-euskarari/manual-de-practicas-ejemplares-en-euskara
http://www.baieuskarari.eus/fr/communaute-bai-euskarari/une-pratique-adequate-et-exemplaire-en-euskaraManual de prácticas ejemplares en euskara / Une pratique adéquate et exemplai...

Manual de prácticas ejemplares en euskara / Une pratique adéquate et exemplai...Bai Euskarari Ziurtagiriaren Elkartea
Destacado (20)
D:\documents and settings\informatica\escritorio\collage ninos indigo[1]![D:\documents and settings\informatica\escritorio\collage ninos indigo[1]](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![D:\documents and settings\informatica\escritorio\collage ninos indigo[1]](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
D:\documents and settings\informatica\escritorio\collage ninos indigo[1]
Manual de prácticas ejemplares en euskara / Une pratique adéquate et exemplai...

Manual de prácticas ejemplares en euskara / Une pratique adéquate et exemplai...
Más de BOSC 2010
Más de BOSC 2010 (20)
Último
💉💊+971581248768>> SAFE AND ORIGINAL ABORTION PILLS FOR SALE IN DUBAI AND ABUDHABI}}+971581248768
+971581248768 Mtp-Kit (500MG) Prices » Dubai [(+971581248768**)] Abortion Pills For Sale In Dubai, UAE, Mifepristone and Misoprostol Tablets Available In Dubai, UAE CONTACT DR.Maya Whatsapp +971581248768 We Have Abortion Pills / Cytotec Tablets /Mifegest Kit Available in Dubai, Sharjah, Abudhabi, Ajman, Alain, Fujairah, Ras Al Khaimah, Umm Al Quwain, UAE, Buy cytotec in Dubai +971581248768''''Abortion Pills near me DUBAI | ABU DHABI|UAE. Price of Misoprostol, Cytotec” +971581248768' Dr.DEEM ''BUY ABORTION PILLS MIFEGEST KIT, MISOPROTONE, CYTOTEC PILLS IN DUBAI, ABU DHABI,UAE'' Contact me now via What's App…… abortion Pills Cytotec also available Oman Qatar Doha Saudi Arabia Bahrain Above all, Cytotec Abortion Pills are Available In Dubai / UAE, you will be very happy to do abortion in Dubai we are providing cytotec 200mg abortion pill in Dubai, UAE. Medication abortion offers an alternative to Surgical Abortion for women in the early weeks of pregnancy. We only offer abortion pills from 1 week-6 Months. We then advise you to use surgery if its beyond 6 months. Our Abu Dhabi, Ajman, Al Ain, Dubai, Fujairah, Ras Al Khaimah (RAK), Sharjah, Umm Al Quwain (UAQ) United Arab Emirates Abortion Clinic provides the safest and most advanced techniques for providing non-surgical, medical and surgical abortion methods for early through late second trimester, including the Abortion By Pill Procedure (RU 486, Mifeprex, Mifepristone, early options French Abortion Pill), Tamoxifen, Methotrexate and Cytotec (Misoprostol). The Abu Dhabi, United Arab Emirates Abortion Clinic performs Same Day Abortion Procedure using medications that are taken on the first day of the office visit and will cause the abortion to occur generally within 4 to 6 hours (as early as 30 minutes) for patients who are 3 to 12 weeks pregnant. When Mifepristone and Misoprostol are used, 50% of patients complete in 4 to 6 hours; 75% to 80% in 12 hours; and 90% in 24 hours. We use a regimen that allows for completion without the need for surgery 99% of the time. All advanced second trimester and late term pregnancies at our Tampa clinic (17 to 24 weeks or greater) can be completed within 24 hours or less 99% of the time without the need surgery. The procedure is completed with minimal to no complications. Our Women's Health Center located in Abu Dhabi, United Arab Emirates, uses the latest medications for medical abortions (RU-486, Mifeprex, Mifegyne, Mifepristone, early options French abortion pill), Methotrexate and Cytotec (Misoprostol). The safety standards of our Abu Dhabi, United Arab Emirates Abortion Doctors remain unparalleled. They consistently maintain the lowest complication rates throughout the nation. Our Physicians and staff are always available to answer questions and care for women in one of the most difficult times in their lives. The decision to have an abortion at the Abortion Cl+971581248768>> SAFE AND ORIGINAL ABORTION PILLS FOR SALE IN DUBAI AND ABUDHA...

+971581248768>> SAFE AND ORIGINAL ABORTION PILLS FOR SALE IN DUBAI AND ABUDHA...?#DUbAI#??##{{(☎️+971_581248768%)**%*]'#abortion pills for sale in dubai@
Último (20)
+971581248768>> SAFE AND ORIGINAL ABORTION PILLS FOR SALE IN DUBAI AND ABUDHA...

+971581248768>> SAFE AND ORIGINAL ABORTION PILLS FOR SALE IN DUBAI AND ABUDHA...
2024: Domino Containers - The Next Step. News from the Domino Container commu...

2024: Domino Containers - The Next Step. News from the Domino Container commu...
Scaling API-first – The story of a global engineering organization

Scaling API-first – The story of a global engineering organization
Handwritten Text Recognition for manuscripts and early printed texts

Handwritten Text Recognition for manuscripts and early printed texts
Tech Trends Report 2024 Future Today Institute.pdf

Tech Trends Report 2024 Future Today Institute.pdf
Axa Assurance Maroc - Insurer Innovation Award 2024

Axa Assurance Maroc - Insurer Innovation Award 2024
Apidays New York 2024 - Scaling API-first by Ian Reasor and Radu Cotescu, Adobe

Apidays New York 2024 - Scaling API-first by Ian Reasor and Radu Cotescu, Adobe
HTML Injection Attacks: Impact and Mitigation Strategies

HTML Injection Attacks: Impact and Mitigation Strategies
Mastering MySQL Database Architecture: Deep Dive into MySQL Shell and MySQL R...

Mastering MySQL Database Architecture: Deep Dive into MySQL Shell and MySQL R...
Boost PC performance: How more available memory can improve productivity

Boost PC performance: How more available memory can improve productivity
From Event to Action: Accelerate Your Decision Making with Real-Time Automation

From Event to Action: Accelerate Your Decision Making with Real-Time Automation
Understanding Discord NSFW Servers A Guide for Responsible Users.pdf

Understanding Discord NSFW Servers A Guide for Responsible Users.pdf
Workshop - Best of Both Worlds_ Combine KG and Vector search for enhanced R...

Workshop - Best of Both Worlds_ Combine KG and Vector search for enhanced R...
TrustArc Webinar - Stay Ahead of US State Data Privacy Law Developments

TrustArc Webinar - Stay Ahead of US State Data Privacy Law Developments
Exploring the Future Potential of AI-Enabled Smartphone Processors

Exploring the Future Potential of AI-Enabled Smartphone Processors
Powerful Google developer tools for immediate impact! (2023-24 C)

Powerful Google developer tools for immediate impact! (2023-24 C)
Puton bosc2010 bio_python-modules-rna
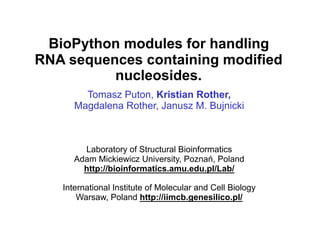
- 1. BioPython modules for handling RNA sequences containing modified nucleosides. Tomasz Puton, Kristian Rother, Magdalena Rother, Janusz M. Bujnicki Laboratory of Structural Bioinformatics Adam Mickiewicz University, Poznań, Poland http://bioinformatics.amu.edu.pl/Lab/ International Institute of Molecular and Cell Biology Warsaw, Poland http://iimcb.genesilico.pl/
- 2. BioPython – a set of freely available tools for biological computation written in Python. http://biopython.org/
- 3. ACUGAUC
- 4. Guanosine (G) ACUGAUC Cytidine (C) Adenosine (A) Uridine (U)
- 5. ACUGAUC >>> from Bio.Alphabet.IUPAC import unambiguous_rna >>> from Bio.Seq import Seq >>> seq = Seq(‘ACUGAUC’, unambiguous_rna)
- 6. ACUGAUC >>> from Bio.Alphabet.IUPAC import unambiguous_rna >>> from Bio.Seq import Seq >>> seq = Seq(‘ACUGAUC’, unambiguous_rna) >>> print seq.reverse_complement() GAUCAGU
- 7. Problem: The linked image cannot be displayed. The file may have been moved, renamed, or deleted. Verify that the link points to the correct file and location. There are 115 known post- transcriptionally modified nucleosides in RNA. Known as: Moreover, several nomenclature wybutosine schemes exist in parallel. yW Y 16G
- 8. Solution: http://github.com/krother/biopython branch rna_alphabet $ git clone git://github.com/krother/biopython.git $ cd biopython $ git checkout rna_alphabet
- 9. Solution: $ python >>> from Bio.Alphabet.ModifiedRNAAlphabet import modified_rna >>> from Bio.RNA.RNASeq import RNASeq
- 10. Solution: $ python >>> from Bio.Alphabet.ModifiedRNAAlphabet import modified_rna >>> from Bio.RNA.RNASeq import RNASeq >>> seq = RNASeq('AG:7CU', modified_rna)
- 11. Solution: $ python >>> from Bio.Alphabet.ModifiedRNAAlphabet import modified_rna >>> from Bio.RNA.RNASeq import RNASeq >>> seq = RNASeq('AG:7CU', modified_rna) >>> print seq[2].full_name 2-O-methyloadenosine
- 12. Solution: $ python >>> from Bio.Alphabet.ModifiedRNAAlphabet import modified_rna >>> from Bio.RNA.RNASeq import RNASeq >>> seq = RNASeq('AG:7CU', modified_rna) >>> print seq[2].full_name 2-O-methyloadenosine >>> print seq[3].long_abbrev m7G
- 14. Example applications (part 1): ModeRNA: A tool for comparative modeling of RNA 3D structure. http://iimcb.genesilico.pl/moderna/ Our software models modified RNA 3D structures! Open source project!
- 15. Example applications (part 2): CompaRNA: A server for continuous benchmarking of automated methods for RNA structure prediction POSTER NUMBER: W17 SUNDAY, JULY 11: 12:40 p.m. – 2.30 p.m. http://comparna.amu.edu.pl/ Uses open source software e.g. BioPython, PyCogent & ModeRNA
- 16. Thank you for attention! Tomasz Puton Kristian Rother t.puton@amu.edu.pl krother@rubor.de Magdalena Rother Janusz M. Bujnicki lenam@amu.edu.pl iamb@genesilico.pl
