Topic 3: Nucleic Acid
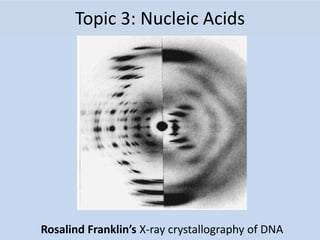
- 1. Topic 3: Nucleic Acids Rosalind Franklin’s X-ray crystallography of DNA
- 2. Essential idea: The structure of DNA is ideally suited to its function 7.1 DNA structure and replication
- 3. 7.1 S.1 Analysis of results of the Hershey and Chase experiment providing evidence that DNA is the genetic material. Hershey and Chase Experiments (1952): Definitive proof that DNA rather than Protein carries the hereditary information of life E. Coli bacteriophage: A virus that infects bacteria. Bacteriophages only contain a protein coat (capsid) and DNA. They wanted to find out whether the protein or DNA carried the genetic instructions to make more viruses. They labeled either the viral proteins or DNA: – Protein capsid: Labeled with radioactive sulfur (35S) – DNA: Labeled with radioactive phosphorus (32P) Radioactive labeled viruses were used to infect cells.
- 4. Either Bacteriophage DNA or Proteins Can be Labeled with Radioactive Elements 7.1 S.1 Analysis of results of the Hershey and Chase experiment providing evidence that DNA is the genetic material.
- 5. Hershey Chase Experiment: DNA is Genetic Material 7.1 S.1 Analysis of results of the Hershey and Chase experiment providing evidence that DNA is the genetic material.
- 6. Hershey and Chase Experiments (1952): Bacterial cells that were infected with the two types of bacteriophage, were then spun down into a pellet (centrifuged), and examined. Results: 1. Labeled viral proteins did not enter infected bacteria (found in supernatant). 2. Labeled viral DNA did enter bacteria during viral infection (found in cell pellet). Conclusion: Protein is not necessary to make new viruses. DNA is the molecule that carries the genetic information to make new viruses!!!! 7.1 S.1 Analysis of results of the Hershey and Chase experiment providing evidence that DNA is the genetic material.
- 7. Rosalind Franklin (1950’s) • Worked with Maurice Wilkins • X-ray crystallography = images of DNA • Provided measurements on chemistry of DNA 7.1 A.1 Rosalind Franklin’s and Maurice Wilkins’ investigation of DNA structure by X-ray diffraction James Watson & Francis Crick (1953) • Discovered the double helix by building models to conform to Franklin’s X-ray data and Chargaff’s Rules.
- 8. DNA Double Helix Nitrogenous Base (A,T,G or C) “Rungs of ladder” “Legs of ladder” Phosphate & Sugar Backbone DNA • Two strands coiled called a double helix • Sides made of a pentose sugar Deoxyribose bonded to phosphate (PO4) groups by phosphodiester bonds • Center made of nitrogen bases bonded together by weak hydrogen bonds 7.1 U.2 DNA structure suggested a mechanism for DNA replication
- 9. DNA • Stands for Deoxyribonucleic acid • Made up of subunits called nucleotides • Nucleotide made of: 1. Phosphate group 2. 5-carbon sugar 3. Nitrogenous base 7.1 U.2 DNA structure suggested a mechanism for DNA replication
- 10. DNA Nucleotide O=P-O O Phosphate Group N Nitrogenous base (A, G, C, or T) CH2 O C1 C4 C3 C2 5 Sugar (deoxyribose) O 7.1 U.2 DNA structure suggested a mechanism for DNA replication
- 11. 11 DNA P P P O O O 1 2 3 4 5 5 3 3 5 P P P O O O 1 2 3 4 5 5 3 5 3 G C T A Hydrogen Bonds
- 12. 7.1 U.1 Nucleosomes help to supercoil the DNA • A nucleosome consists of DNA wrapped around 8 histone proteins (prokaryotic cells lack these proteins making there DNA “naked). • The DNA wraps twice around the histone protein core. • Another histone protein is attached to the outside of the DNA strand. It helps maintain the colloidal structure of the nucleosome. • DNA, because of its negative charge is attracted to the positive charge on the amino acids of the histone proteins. • Tails of neighboring histones, link up during chromosomal condensation, causing the nucleosomes to pull closer together. • This is part of the supercoiling process that occurs during mitosis and meiosis • Supercoiling in general helps regulate transcription because only certain areas of the DNA are accessible for the production of mRNA by transcription. This regulates the production of a polypeptide. http://pbil.univlyon1.fr/members/sagot/htdocs/tea m/projects/chromo_net/images/epi.jpg
- 14. DNA Replication 7.1 U.3 DNA polymerases can only add nucleotides to the 3’ end of a primer
- 15. Synthesis Phase (S phase) • S phase during interphase of the cell cycle • Nucleus of eukaryotes Mitosis -prophase -metaphase -anaphase -telophase G1 G2 S phase interphase DNA replication takes place in the S phase. Mitosis -prophase -metaphase -anaphase -telophase G1 G2 S phase interphase DNA replication takes place in the S phase. 7.1 U.3 DNA polymerases can only add nucleotides to the 3’ end of a primer Super coiling begin in prophase, making chromosomes visible for the first time
- 16. 2.7 S.2 Analysis of Meselson and Stahl’s results to obtain support for the theory of semi- conservative replication of DNA. https://upl oad.wikimedia.org/wikipedia/commons/a/a2/DNAreplicationModes.png Before Meselson and Stahl’s work there were different proposed models for DNA replication. After their work only semi-conservative replication was found to be biologically significant.
- 17. 2.7 S.2 Analysis of Meselson and Stahl’s results to obtain support for the theory of semi- conservative replication of DNA. Learn about Meselson and Stahl’s work with DNA to discover the mechanism of semi-conservative replication http://highered.mheducation.com/olcweb/cgi/pluginpop.cg i?it=swf::535::535::/sites/dl/free/0072437316/120076/bio2 2.swf::Meselson%20and%20Stahl%20Experiment http://www.nature.com/scitable/topicpage/Semi-Conservative-DNA-Replication-Meselson-and-Stahl-421#
- 18. • DNA replication is very specific to the arrangements of base pairs • In DNA replication, the strands separate – Enzymes use each strand as a template to assemble the new strands DNA REPLICATION Parental molecule of DNA Both parental strands serve as templates Two identical daughter molecules of DNA Nucleotides A 7.1 U.3 DNA polymerases can only add nucleotides to the 3’ end of a primer
- 19. 1. Helicase: unwinds DNA at origins of replication 2. Initiation proteins separate 2 strands forms replication bubble 3. Single-Strand Binding Proteins attach and keep the 2 DNA strands separated and untwisted 4. Primase: puts down RNA primer to start replication 5. DNA polymerase III: adds complimentary bases to leading strand (new DNA is made 5’ 3’) 6. Lagging strand grows in 3’5’ direction by the addition of Okazaki fragments 7. DNA polymerase I: replaces RNA primers with DNA 8. DNA ligase: seals fragments together 9. DNA gyrase: an enzyme that relieves strain while double-strand DNA is being unwound by helicase. 7.1 U.5 DNA replication is carried out by a complex system of enzymes. [The proteins and enzymes involved in DNA replication should include helicase, DNA gyrase, single strand binding proteins, DNA primase and DNA polymerases I and III.] Major Steps of DNA Replication:
- 20. DNA Gyrase SSBP DNA Poly Lead Strand Lagging Strand DNA Ligase RNA Primer Helicase Replication Direction RNA Primase
- 21. Helicase The ‘ase’ ending indicates it is an enzyme. Helicase is DNA’s origin of replication and creates replication forks 2.7 U.2 Helicase unwinds the double helix and separates the two strands by breaking hydrogen bonds.
- 22. • DNA replication begins at the different origins in the 5’ to 3’ direction at the replication fork. • RNA primase (is the primer to that starts the process) attaches to the DNA and adds a small RNA primer to provide a free 3’ OH starting point since DNA polymerases can only add nucleotides to the 3’ end of a primer • DNA polymerase III adds free nucleotides in the 5’ to 3’ direction in the direction of the replication fork. 7.1 U.3 DNA polymerases can only add nucleotides to the 3’ end of a primer
- 23. • Begins at Origins of Replication • Two strands open forming Replication Forks (Y- shaped region) • New strands grow at the forks • The leading strand (copies in one long continuous piece) begins on the 3’ side and the lagging strand begins on the 5’ side (copies in small fragments, which must be pieced together) Replication Fork Parental DNA Molecule 3’ 5’ 3’ 5’ 7.1 U.4 DNA replication is continuous on the leading strand and discontinuous on the lagging strand. [Details of DNA replication differ between prokaryotes and eukaryotes. Only the prokaryotic system is expected.]
- 24. Remember HOW the Carbons Are Numbered! O O=P-O O Phosphate Group N Nitrogenous base (A, G, C, or T) CH2 O C1 C4 C3 C2 5 Sugar (deoxyribose)
- 25. Leading strand vs. Lagging strand
- 26. Primase adds RNA primer is RNA that initiates DNA synthesis. DNA Gyrase 7.1 U.3 DNA polymerases can only add nucleotides to the 3’ end of a primer
- 27. 2.7 U.3 DNA polymerase links nucleotides together to form a new strand, using the pre- existing strand as a template. DNA polymerase always moves in a 5’ to 3’ direction • DNA polymerase catalyzes the covalent phosphodiester bonds between sugars and phosphate groups forming covalent bonds
- 28. DNA polymerase III adds nucleotides in 5’3’ direction on leading strand
- 29. 2.7 U.1 The replication of DNA is semi-conservative and depends on complementary base pairing. https://upload.wikimedia.org/wikipedia/commons/3/33/DNA_replication_split_horizontal.svg 1. Each of the nitrogenous bases can only pair with its partner (A=T and G=C) this is called complementary base pairing. 2. The two new strands formed will be identical to the original strand.
- 30. 2.7.U1 The replication of DNA is semi-conservative and depends on complementary base pairing. https://upload.wikimedia.org/wikipedia/commons/3/33/DNA_replication_split_horizontal.svg 3. Each new strand contains one original and one new strand, therefore DNA Replication is said to be a Semi- Conservative Process.
- 31. Replication on leading strand 1. RNA Primase attaches RNA Primer 2. DNA Polymerase attaches nucleotides in a 5’ to 3’ direction 7.1 U.4 DNA replication is continuous on the leading strand and discontinuous on the lagging strand. [Details of DNA replication differ between prokaryotes and eukaryotes. Only the prokaryotic system is expected.]
- 32. Replication on lagging strand 1. RNA Primase attaches multiple pieces of RNA Primer 2. DNA Polymerase attaches nucleotides in a 5’ to 3’ direction in between the Primer pieces creating Okazaki fragments 3. DNA Polymerase replaces the RNA primer pieces with DNA 4. Ligase glues the fragments together
- 33. 1. Helicase: unwinds DNA at origins of replication Initiation proteins separate 2 strands forms replication bubble 2. Single-Strand Binding Proteins attach and keep the 2 DNA strands separated and untwisted 3. RNA Primase: puts down RNA primer to start replication 4. DNA polymerase III: adds complimentary bases to leading strand (new DNA is made 5’ 3’) 5. Lagging strand grows in 3’5’ direction by the addition of Okazaki fragments 6. DNA polymerase I: replaces RNA primers with DNA 7. DNA ligase: seals fragments together 8. DNA gyrase: an enzyme that relieves strain while double-strand DNA is being unwound by helicase. 7.1 U.5 DNA replication is carried out by a complex system of enzymes. [The proteins and enzymes involved in DNA replication should include helicase, DNA gyrase, single strand binding proteins, DNA primase and DNA polymerases I and III.] Major Steps of DNA Replication:
- 34. DNA Gyrase SSBP DNA Poly Lead Strand Lagging Strand DNA Ligase RNA Primer Helicase Replication Direction RNA Primase
- 35. • Dideoxyribonucleotides inhibit DNA polymerase during replication, thereby stopping replication from continuing. • Dideoxyribonucleotides with fluorescent markers, is incorporated into sequences of DNA, to stop replication at the point at which they are added. • This creates different sized fragments with fluorescent markers that can be separated by gel electrophoresis and analyzed by comparing the color of the fluorescence with the fragment length. 7.1 A.2 Use of nucleotides containing deoxyribonucleic acid to stop DNA replication in preparation of samples for base sequencing
- 36. Problem at the 5’ End • DNA poly only adds nucleotides to 3’ end • No way to complete 5’ ends of daughter strands • Over many replications, DNA strands will grow shorter and shorter 7.1 U.6 Some regions of DNA do not code for proteins but have other important functions. [The regions of DNA that do not code for proteins should be limited to regulators of gene expression, introns, telomeres and genes for tRNAs.]
- 37. Telomeres: repeated units of short nucleotide sequences (TTAGGG) at ends of DNA • Telomeres “cap” ends of DNA to postpone erosion of genes at ends (TTAGGG) • Telomerase: enzyme that adds to telomeres – Eukaryotic germ cells, cancer cells Telomeres stained orange at the ends of mouse chromosomes 7.1 U.6 Some regions of DNA do not code for proteins but have other important functions. [The regions of DNA that do not code for proteins should be limited to regulators of gene expression, introns, telomeres and genes for tRNAs.]
- 38. Telomeres & Telomerase 7.1 U.6 Some regions of DNA do not code for proteins but have other important functions. [The regions of DNA that do not code for proteins should be limited to regulators of gene expression, introns, telomeres and genes for tRNAs.]
- 39. 1.6 Cell division Essential idea: Cell division is essential but must be controlled.
- 40. Why do cells divide: • Growth: Multicellular organisms increase their size by increasing their number of cells through mitosis • Asexual reproduction: Certain eukaryotic organisms may reproduce asexually by mitosis (e.g. vegetative reproduction) • Tissue Repair: Damaged tissue can recover by replacing dead or damaged cells • Embryonic development: A fertilized egg (zygote) will undergo mitosis and differentiation in order to develop into an embryo
- 41. • Cellular division in eukaryotic cells. • Chromatin is arranged into chromosomes. • Chromosomes double. • Cell grows in size. • Cells divide. • Is cellular cloning. Cell division
- 42. 2 phases: 1. Interphase 2. M phase (mitotic phase) a. Prophase b. Metaphase c. Anaphase d. Telophase & cytokinesis Figure 12.4 The cell cycle Phases of the Cell Cycle (life cycle of a cell)
- 43. Interphase • The non-dividing phase in a cell • Lasts about ~ 90% of the cell cycle. • The cell grows and replicates DNA preparing for Mitosis. • There are three periods: 3 periods of Interphase 1. Go – a cell functioning as normal 2. G1 phase – first growth phase 3. S phase- synthesis of DNA 4. G2 phase- 2nd growth phase Mitosis is a reliable process. Only one error occurs per 100,000 cell divisions. 1.6 U.4 Interphase is a very active phase of the cell cycle with many processes occurring in the nucleus and cytoplasm.
- 44. 1.6 U.4 Interphase is a very active phase of the cell cycle with many processes occurring in the nucleus and cytoplasm. Interphase This when the cell carries out it’s normal functions Metabolic reactions (e.g. respiration to produce ATP) are necessary for the life of the cell Protein synthesis - proteins and enzymes are necessary to allow cell grow Organelles numbers are increased to first support the enlarged cell DNA is replicated to ensure a second copy is available to enable mitosis Cells spend the majority of their time in interphase. It is a very active phase of the cycle. Mr P O D http://botit.botany.wisc.edu/Resources/Botany/Mitosis/Allium/Various%20views/Interphase%20prophase.JPG
- 45. 1.6 U.5 Cyclins are involved in the control of the cell cycle. Cyclinsare a family of proteins that control the progression of cells through the cell cycle Cells cannot progress to the next stage of the cell cycle unless the specific cyclin reaches it threshold. http://upload.wikimedia.org/wikipedia/commons/thumb/9/99/Protein_CCNE1_PDB_1w98.png/800px-Protein_CCNE1_PDB_1w98.png Cyclins bind to enzymes called cyclin-dependent kinases These kinases then become active and attach phosphate groups to other proteins in the cell. The attachment of phosphate triggers the other proteins to become active and carry out tasks (specific to one of the phases of the cell cycle). 4 3 2 1
- 46. http://upload.wikimedia.org/wikipedia/commons/thumb/9/99/Protein_CCNE1_PDB_1w98.png/800px-Protein_CCNE1_PDB_1w98.png Triggers cells to move from G0 to G1 and from G1 into S phase. prepares the cell for DNA replication in S phase. activates DNA replication inside the nucleus in S phase. promotes the assembly of the mitotic spindle and other tasks in the cytoplasm to prepare for mitosis. Progression through parts of the cell cycle are affected in various ways by specific cyclins
- 47. 1.6 U.1 Mitosis is division of the nucleus into two genetically identical daughter nuclei. http://commons.wikimedia.org/wiki/File:Chromosome.svg centromere is the part of a chromosome that links sister chromatids Sister chromatids are duplicated chromosomes attached by a centromere Get the terminology right centrioles organize spindle microtubules Spindle microtubules (also referred to as spindle fibers) In animal cells two centrioles are held by a protein mass referred to as a centrosome After anaphase when the sister chromatids separate they should then be referred to as chromosomes It is easy to misuse the terms chromatid and chromosome. It is even easier to confuse the terms centromere, centriole and centrosome due to their similar spelling. Keep the terms clear in your mind to avoid losing marks. http://commons.wikimedia.org/wiki/Mitosis#mediaviewer/File:Mitosis_cells_sequence.svg
- 48. 1.6 U.2 Chromosomes condense by supercoiling during mitosis. Why supercoil chromosomes? Human cells are on average 10μm in diameter and the nucelus within each is less than 5 μm in diameter. Human chromosomes are 15mm to 85mm (15,000μm to 85,000 μm) in length. Chromosomes need to be stored compactly to fit within the nuclei of cells. This problem becomes more acute during mitosis when chromosomes need to be short and compact enough that they can be separated and moved to each end of the cell.
- 49. 1.6 U.2 Chromosomes condense by supercoiling during mitosis. How are chromosomes supercoiled? Strain is placed on a DNA helix by over winding or under winding of the helix This causes the DNA molecule to coil back on itself becoming shorter and wider Remember that in eukaryotes proteins called histones form nucleosomes to aid the process of supercoiling http://www.maths.uq.edu.au/~infinity/Infinity7/images/supercoiling.gifhttp://vanat.cvm.umn.edu/mMeiosis/images/chromosome-X.jpg
- 50. http://highered.mheducation.com/sites/0072495 855/student_view0/chapter2/animation__mitosis _and_cytokinesis.html Use the animated tutorials to learn about mitosis http://www.johnkyrk.com/mitosis.html http://www.sumanasinc.com/webcontent/animations/content /mitosis.html http://outreach.mcb.harvard.edu/animations/cellcycle. swf
- 51. Prophase • The nucleolus disappears. • Chromatin condenses into visible chromosomes. • There are two sister chromatids held together by a centromere. • The mitotic spindle forms in the cytoplasm. . 1.6 S.1 Identification of phases of mitosis in cells viewed with a microscope or in a micrograph
- 52. Metaphase • The nuclear envelope disappears. • Spindle fibers extend from each pole to the cell’s equator. • Spindle fibers attach to the centromeres.
- 53. Figure 12.3 Chromosome duplication and distribution during mitosis
- 54. Anaphase • Characterized by movement. It begins when pairs of sister chromatids pull apart. • Sister chromatids move to opposite poles of the cell. • Chromosomes look like a “V” as they are pulled. • At the end of anaphase, the two poles have identical number and types of chromosomes.
- 55. Telophase • Microtubules elongate the cell. • Daughter nuclei begin to form at the two poles. • Nuclear envelopes re-form. • Nucleolus reappears. • Chromatin uncoils. • The cells cytoplasm begins to pinch. • It is basically the opposite of prophase.
- 56. 1.6 U.3 Cytokinesis occurs after mitosis and is different in plant and animal cells. mitosis is the division of the nucleus, cytokinesis is the division of the cytoplasm to create two cells Though mitosis is similar for animal and plant cells cytokinesis is very different. http://wwwprod.biochem.wisc.edu/biochem/faculty/bednarek/images/figure_color.gif http://glencoe.mheducation.com/sites/983 4092339/student_view0/chapter10/animati on_-_cytokinesis.html http://www.haroldsmithlab.com/images/pg_HeLa_cell_division.jpg
- 57. Figure 12.8 Cytokinesis in animal and plant cells
- 58. 1.6 S.1 Identification of phases of mitosis in cells viewed with a microscope or in a micrograph. 1.6 S.2 Determination of a mitotic index from a micrograph. http://www.nuffieldfoundation.org/practical-biology/investigating-mitosis-allium-root-tip-squash A very good, well explained lab outline for creating slides and calculating the mitotic index. http://www.biology.arizona.edu/cell_bio/activities/cell_cycle/cell_cycle.html An excellent online alternative if resources don’t permit students to create and view their own slides
- 59. 1.6 U.6 Mutagens, oncogenes and metastasis are involved in the development of primary and secondary tumors. Tumors are abnormal growth of tissue that develop at any stage of life in any part of the body. A cancer is a malignant tumour and is named after the part of the body where the cancer (primary tumour) first develops. Use the links to find out: • most common types of cancer • what causes cancer and associated risk factors • how cancer can be treated
- 60. 1.6 U.6 Mutagens, oncogenes and metastasis are involved in the development of primary and secondary tumors. mutation in a oncogene If a mutation occurs in an oncogenes it can become cancerous. In normal cells oncogenes control of the cell cycle and cell division. http://en.wikipedia.org/wiki/Oncogene#mediaviewer/File:Oncogenes_illustration.jpg uncontrolled cell division tumor formation malfunction in the control of the cell cycle
- 61. 1.6 U.6 Mutagens, oncogenes and metastasis are involved in the development of primary and secondary tumors. Mutagens are agents that cause gene mutations. Not all mutations result in cancers, but anything that causes a mutation has the potential to cause a cancer. Mutagens can be: • chemicals that cause mutations are referred to as carcinogens • high energy radiation such as X-rays • short-wave ultraviolet light • Some viruses A mutation is a change in an organisms genetic code. A mutation/change in the base sequence of a certain genes can result in cancer. http://en.wikipedia.org/wiki/Oncogene#mediaviewer/File:Oncogenes_illustration.jpg
- 62. 1.6 U.6 Mutagens, oncogenes and metastasis are involved in the development of primary and secondary tumors. Factors (other than exposure to mutagens) that increase the probability of tumour development include: • The vast number of cells in a human body – the greater the number of cells the greater the chance of a mutation. • The longer a life span the greater the chance of a mutation. Several mutations must occur in the same cell for it to become a tumour causing cell. The probability of this happening in a single cell is extremely small. http://en.wikipedia.org/wiki/Oncogene#mediaviewer/File:Oncogenes_illustration.jpg
- 63. 1.6 A.1 The correlation between smoking and incidence of cancers. http://en.wikipedia.org/wiki/File:Smoking_lung_cancer.png There are many other similar surveys in different countries, with different demographics that show similar results. Along with lung cancer, cancers of mouth and throat are very common as these areas are in direct contact with the smoke too. It might surprise you that the following cancers are also more common in smokers: • Head and neck • Bladder • Kidneys • Breast • Pancreas • Colon
- 64. a. Describe the relationship shown. b. What type of correlation is shown c. How strong is the correlation? Justify your answer by discussing the evidence. d. The correlation shown here is lagged. A lag is a time gap between the factors. Estimate the size of the lag between cigarette consumption and lung cancer death.
- 65. http://en.wikipedia.org/wiki/File:Smoking_lung_cancer.png a. Describe the relationship shown. b. What type of correlation is shown c. How strong is the correlation? Justify your answer by discussing the evidence. d. The correlation shown here is lagged. A lag is a time gap between the factors. Estimate the size of the lag between cigarette consumption and lung cancer death. There are many other similar surveys in different countries, with different demographics that show similar results. Along with lung cancer, cancers of mouth and throat are very common as these areas are in direct contact with the smoke too. It might surprise you that the following cancers are also more common in smokers: • Head and neck • Bladder • Kidneys • Breast • Pancreas • Colon
- 66. Essential idea: Information stored as a code in DNA is copied onto mRNA 7.2 Transcription & Gene Expression http://www.knowingforsure.com/wp-content/uploads/2015/01/Traits.jpg Trait vs Fate
- 67. 2.7 U.4 Transcription is the synthesis of mRNA copied from the DNA base sequences by RNA polymerase. 2.7 U.5 Translation is the synthesis of polypeptides on ribosomes. Q - What is the purpose of transcription and translation? A - Gene expression the processes of create a polypeptides which in turns folds to become a protein. Proteins carry out many essential functions in cells. For more detail review 2.4.U7. Rhodopsin - A Light absorbing pigment
- 68. Rubisco • Full name ribulose bisphosphate carboxylase • Enzyme - catalyzes the reaction that fixes carbon dioxide from the atmosphere • Provides the source of carbon from which all carbon compounds, required by living organisms, are produced. • Found in high concentrations in leaves and algal cells http://upload.wikimedia.org/wikipedia/commons/b/b0/Mint-leaves-2007.jpg 2.4 A.1 Rubisco, insulin, immunoglobulins, rhodopsin, collagen and spider silk as examples of the range of protein functions.
- 69. Collagen • A number of different forms • All are rope-like proteins made of three polypeptides wound together. • About a quarter of all protein in the human body is collagen • Forms a mesh of fibers in skin and in blood vessel walls that resists tearing. • Gives strength to tendons, ligaments, skin and blood vessel walls. • Forms part of teeth and bones, helps to prevent cracks and fractures to bones and teeth https://en.wikipedia.org/wiki/Tooth_(human)#med iaviewer/File:Teeth_by_David_Shankbone.jpg 2.4 A.1 Rubisco, insulin, immunoglobulins, rhodopsin, collagen and spider silk as examples of the range of protein functions.
- 70. spider silk • Different types of silk with different functions • Dragline silk is stronger than steel and tougher than Kevlar • When first made it contains regions where the polypeptide forms parallel arrays (bottom) • Some regions seem like a disordered tangle (middle) • When the stretched the polypeptide gradually extends, making the silk extensible and very resistant to breaking. https://en.wikipedia.org/wiki/Spider_silk#mediaviewer/File:Structure_of_spider_silk_thread_Modified.svg 2.4 A.1 Rubisco, insulin, immunoglobulins, rhodopsin, collagen and spider silk as examples of the range of protein functions.
- 71. 2.7 U.4 Transcription is the synthesis of mRNA copied from the DNA base sequences by RNA polymerase 2.7 U.5 Translation is the synthesis of polypeptides on ribosomes. http://learn.genetics.utah.edu/content/molecules/transcribe/
- 72. 7.1 U.1 Nucleosomes help to supercoil the DNA. 7.2 U.5 Gene expression is regulated by proteins that bind to specific base sequences in DNA. • A nucleosome consists of DNA wrapped around 8 histone proteins (prokaryotic cells lack these proteins making there DNA “naked). • The DNA wraps twice around the histone protein core. • Another histone protein is attached to the outside of the DNA strand. It helps maintain the colloidal structure of the nucleosome. • DNA, because of its negative charge is attracted to the positive charge on the amino acids of the histone proteins. • Tails of neighboring histones, link up during chromosomal condensation, causing the nucleosomes to pull closer together. • This is part of the supercoiling process that occurs during mitosis and meiosis • Supercoiling in general helps regulate transcription because only certain areas of the DNA are accessible for the production of mRNA by transcription. This regulates the production of a polypeptide. http://pbil.univlyon1.fr/members/sagot/htdocs/tea m/projects/chromo_net/images/epi.jpg
- 74. 7.2 U.2 Nucleosomes help to regulate transcription in eukaryotes. • Supercoiling helps regulate transcription, one supercoiling modification is through the modification of the histone tails. • Acetylation Acetyl groups can be added to the positively charged histone tails, they become negative and that repels the negatively charged DNA. This opens up the nucleosome so the DNA is not as close to the histone anymore causing gene expression • Methylation Methyl group is non polar which causes DNA remains tightly packed and transcription is inhibited.
- 75. 7.2 S.1 Analysis of changes in the DNA methylation patterns. http://i.dailymail.co.uk/i/pix/2008/09/12/article-1054890- 029CF17900000578-854_233x364.jpg • Another way gene expression can be controlled is through methylation (adding a methyl CH3 group) to the histone proteins. • Methylation of the histone proteins inhibites transcription of the gene • The amount of methylation can vary over an organisms lifetime and can be affected by environmental factors
- 76. 7.2 U.6 The environment of a cell and of an organism has an impact on gene expression. The impact gene expression Morphogenic Effect (Aging affects) the accumulation of damage cells over a lifetime; decreases the capacity to maintain homeostasis
- 77. 7.2 U.2 Nucleosomes help to regulate transcription in eukaryotes. Epigenetics • The changes related to gene expression or cellular phenotype of without changes to the nucleotide sequence of the genome. • Examples of mechanisms that produce such changes are DNA methylation and histone modification of the nucleosomes, each of which alters how genes are expressed without altering the underlying DNA sequence. Trait vs Fate
- 78. 2.7.U4 Transcription is the synthesis of mRNA copied from the DNA base sequences by RNA polymerase. Step one: Transcription the process by which an RNA sequence is produced from a DNA template: Gene expression is the constructions of a protein from the DNA using RNA. RNA with the help of ribosomes constructs a protein from amino acids
- 79. 7.2 A.1 The promoter as an example of non-coding DNA with a function. • The promoter region is a DNA sequence that initiates transcription and is an example of non- coding DNA that plays a role in gene expression. This promoter region is called the TATA box. • The promoter sequence is located near the start site of transcription and is where the RNA polymerase binds in order for transcription to take place. • DNA always is copied in a 5’ to 3’ direction. http://study.com/cimages/multimages/16/junk_dna_1.jpg
- 80. • The enzyme RNA polymerase binds to a site on the DNA at the start of a gene (The sequence of DNA that is transcribed into RNA is called a gene). • RNA polymerase separates the DNA strands and synthesizes a complementary RNA copy from the antisense DNA strand. 2.7 U.4 Transcription is the synthesis of mRNA copied from the DNA base sequences by RNA polymerase.
- 81. 7.2 U.3 Eukaryotic cells modify mRNA after transcription. http://i.dailymail.co.uk/i/pix/2008/09/12/article-1054890- 029CF17900000578-854_233x364.jpg a) The gene has a promotor region and a terminator region b) Transcription requires the presence of a regulator protein from another gene (possible from another chromosome). c) The RNA polymerase can now bind to the promotor and begin the transcription of the gene. d) The mRNA is transcribed including introns e) The completed mRNA which will require post transcriptional modification to remove the introns. click4biology TATA box
- 82. 7.2 U.1 Transcription occurs in a 5’ to 3’ direction. [RNA polymerase adds the 5´ end of the free RNA nucleotide to the 3´ end of the growing mRNA molecule.] • Transcription occurs in a 5’ to 3’ direction where the 5’ end of the free RNA nucleotide is added to the 3’ end of the RNA molecule that is being synthesized. Consisting of 3 stages called 1. Initiation RNA polymerase binds to the promoter with the help of specific binding proteins 2. Elongation transcription machinery needs to move histones out of the way, unwinding DNA, allowing RNA Polymerase to synthesis of a new RNA strand in the 5′ to 3′ direction 3. Termination RNA synthesis will continue along the DNA template strand until the polymerase encounters a signal that tells it to stop
- 83. 7.1 U.6 Some regions of DNA do not code for proteins but have other important functions. [The regions of DNA that do not code for proteins should be limited to regulators of gene expression, introns, telomeres and genes for tRNAs.] • There are many areas of DNA containing repetitive sequences, especially in eukaryotic DNA, in humans it makes up between 24-37% of our genome. These repetitive areas usually occurs near the ends of chromosomes. • Introns are non-coded regions, exons are coded areas for a protein, these two areas must be separated and introns must be removed before leaving the nucleus for protein synthesis to take place. *Spliceosome are enzymes constructed from RNA and are used in eukaryotic cell to remove these introns. http://www.phschool.com/science/biology_place/bio coach/images/transcription/eusplice.gif
- 84. Post Transcriptional Modification Pre mRNA Mature mRNA
- 85. 7.1 U.6 Some regions of DNA do not code for proteins but have other important functions. [The regions of DNA that do not code for proteins should be limited to regulators of gene expression, introns, telomeres and genes for tRNAs.] • In addition to introns, some repetitive sequences are called telomeres, these areas protect DNA during replication. Telomeres are the caps at the end of each strand of DNA that protect our chromosomes, like the plastic tips at the end of shoelace They prevents the loss of genes near the end of the chromosomes. Telomeres stained green and red at the ends of the chromosomes http://www.newswise.com/images/uplo ads/2010/09/28/karlsederhr.jpg
- 86. 7.2 U.4 Splicing of mRNA increases the number of different proteins an organism can produce 1. Promotor region 2. Free Nucleotide Phosphates 3. Addition of Nucleotides to the new mRNA 4. Early mRNA 5. Early mRNA showing introns (non-coding) 6.Introns removed by the enzyme spliceosome allowing exons to combine 7. Mature mRNA ready for translation 8. mRNA going to cytoplasm. click4biology
- 87. 7.2 U.4 Splicing of mRNA increases the number of different proteins an organism can produce. https://commons.wikimedia.org/wiki/File:DNA_alternative_splicing.gif The splicing process above can happen in different ways to the same gene, particular exons (of a gene) may be included within or excluded from mature mRNA Multiple proteins produced by a single gene. Each proteins produced will vary in it’s biological function. An example of this is the IgM gene which produces different immunoglobulins (antibodies) to fight different pathogens.
- 88. • The variable nature of the Short tandem repeats (STR) regions that are analyzed for forensic testing intensifies the discrimination between one DNA profile and another. Theses sections have high rates of mutations and change frequently. Forensic science takes advantage of the population's variability in STR lengths, enabling scientists to distinguish one DNA sample from another. For example, the likelihood that any two individuals (except identical twins) will have the same 13-loci DNA profile can be as low as 1 in 1 billion or less. 7.1 A.3 Tandem repeats are used in DNA profiling.
- 89. 7.1 A.3 Tandem repeats are used in DNA profiling. • Short tandem repeats (STRs), also known as variable tandem repeats (VNTRs) are regions of noncoding DNA that contain repeats of the same nucleotide sequence. These short repeats show variations between individuals in terms of the number of times the sequences is repeated. Example • CATACATACATACATACATACATACATA repeated 7 times for one individual. However, in another individual, 11 times CATACATACATACATACATACATACATACATA CATACATACATA. • Used in DNA profiling used in crime scene investigations, genealogical and paternity tests.
- 90. 7.2 U.6 The environment of a cell and of an organism has an impact on gene expression. • The environment, as well as the organism's internal world, which includes such factors as its hormones and metabolism can have an impact on gene expression • Temperature and light are external conditions which can affect gene expression in certain organisms. • As an example, Himalayan rabbits carry the gene, which is required for the development of pigments in the fur, skin, and eyes, and whose expression is regulated by temperature • Specifically, a gene called the C gene is inactive above 35°C, and it is maximally active from 15°C to 25°C. This temperature regulation of gene expression produces rabbits with a distinctive coat coloring. • In the warm weather no pigments fur is white • In low temperature the rabbit's extremities (i.e., the ears, tip of the nose, and feet), where the, the C gene actively produces pigment, making these parts of the animal black. http://upload.wikimedia.org/wikipedia/commons/0/06/Kr%C 3%B3liki_kalifornijskie_666.jpg http://upload.wikimedia.org/wikipedia/en/8/81/Kostya2.jpg
- 91. Essential idea: Information transferred from DNA to mRNA is translated into an amino acid sequence. 7.3 Translation Section of Titin, our largest known protein http://circ.ahajournals.org/content/124/8/876/F2.large.jpg
- 92. Components of Translation 1. mRNA = message 2. tRNA = interpreter 3. Ribosome = site of translation 7.3 U.1 Initiation of translation involves assembly of the components that carry out the process.
- 93. 2.7 U.5 Translation is the synthesis of polypeptides on ribosomes. http://www.nature.com/scitable/topicpage/ribosomes-transcription-and-translation-14120660 Translation is the process of protein synthesis in which the genetic information encoded in mRNA is translated into a sequence of amino acids in a polypeptide chain A ribosome is composed of two halves, a large and a small subunit. During translation, ribosomal subunits assemble together like a sandwich on the strand of mRNA: • Each subunit is composed of RNA molecules and proteins • The small subunit binds to the mRNA • The large subunit has binding sites for tRNAs and also catalyzes peptide bonds between amino acids
- 94. Ribosomes Active sites: • A site: holds AA to be added • P site: holds growing polypeptide chain • E site: exit site for tRNA 7.3 U.1 Initiation of translation involves assembly of the components that carry out the process.
- 95. 7.3 S.2 The use of molecular visualization software to analyse the structure of eukaryotic ribosomes and a tRNA molecule. D-Loop Ribosome sight recognition T-Loop Ribosome sight recognition Acceptor End With the help of ATP this is the site of attachment of The amino acid
- 96. 7.3 S.2 The use of molecular visualization software to analyse the structure of eukaryotic ribosomes and a tRNA molecule. D-Loop Ribosome sight recognition T-Loop Ribosome sight recognition Acceptor End With the help of ATP this is the site of attachment of The amino acid
- 97. 7.3 A.1 tRNA activating enzymes illustrate enzyme–substrate specificity and the role of phosphorylation. Building tRNA binds with a specific amino acid is a catalyzed reaction 1. tRNA-activating enzyme 2. ATP binds to the enzyme. 3. Specific amino acid binds to the acceptor site(ACC) on the tRNA molecule. Building tRNA binds with a specific amino acid
- 98. 7.3 A.1 tRNA-activating enzymes illustrate enzyme–substrate specificity and the role of phosphorylation. http://www.phschool.com/science/biology_place/biocoach/translation/addani.html
- 99. tRNA • Transcribed in nucleus • Specific to each amino acid • Transfer Amino Acids to ribosomes • Anticodon: pairs with complementary mRNA codon • Base-pairing rules between 3rd base of codon & anticodon are not as strict. 7.3 U.1 Initiation of translation involves assembly of the components that carry out the process.
- 100. Translation stages: Initiation, Elongation and Termination • Translation occurs in the 5' to 3' direction along the mRNA A. Initiation begins with the attachment of the ribosome to the mRNA using the start codon AUG B. Elongation The ribosome moves one codon along the mRNA (in a 5’ – 3’ direction): • The tRNA in the P site is moved to the E site and then released • The tRNA in the A site is moved into P site C. Termination occurs at the STOP codon (UGA, UAG or UAA). • a release factor attaches to the A site • the polypeptide chain is released 7.3 U.1 Initiation of translation involves assembly of the components that carry out the process.
- 101. The role of RNA in Protein Synthesis • 3 Types of RNA molecules in the steps from gene to protein: 1. Messenger RNA (mRNA), Makes a complimentary copy of DNA in the form of RNA. Length varies depending on the gene sequence 2. Transfer RNA (tRNA) carries amino acid to the site of synthesis. 3. Ribosomal RNA (rRNA), stabilizes the site of synthesis 7.3 U.1 Initiation of translation involves assembly of the components that carry out the process.
- 102. 7.3 U.2 Synthesis of the polypeptide involves a repeated cycle of events. • Step one of translation is the small and large sub units of the ribosome come together between the mRNA sequence • Step two tRNA (carrying methionine (Met), the start code) attaches to the mRNA at the A site • Step three the first tRNA moves to the P, a second tRNA located at the A. The two amino acids form a peptide bond.
- 103. 7.3 U.2 Synthesis of the polypeptide involves a repeated cycle of events. • The two amino acids are joined together through a condensation reaction that creates a peptide bond between the two amino acids. • Step Four The ribosome moves along the mRNA one codon shifting the tRNA that was attached to methionine to the E site. • The tRNA is released back into the cytoplasm from the E site, allowing it to pick up another amino acid (methionine) to build another polypeptide.
- 104. 7.3 U.2 Synthesis of the polypeptide involves a repeated cycle of events. • Another tRNA moves into the empty A site bringing the next amino acid corresponding to themRNA codon. • Again, the amino acid is attached to the polypeptide forming a peptide bond, the ribosome slides across one codon and tRNA at the P site moves into the E site releasing it back into the cytoplasm. • The ribosome continues to move along the mRNA adding amino acids to the polypeptide chain. • This process continues until a stop codon is reached.
- 105. 7.3 U.3 Disassembly of the components follows termination of translation. • Termination begins when 1 of the 3 stop codons UAA UGA UAG moves into the A site. • These tRNA have no attached amino acids. • When the stop codon is reached the ribosome dissociates and the polypeptide is released.
- 106. https://www.youtube.com/watch?v=G2yovIdpTVk Watch the animation about the process of translation.
- 108. 2.7 U.6 The amino acid sequence of polypeptides is determined by mRNA according to the genetic code. The central dogma of genetics Messenger RNA (mRNA): A transcript copy of a gene used to encode a polypeptide • The length of mRNA molecules varies – 23,000 different genes, the average length for mammals is approximately 2,200 nucleotides (this translates to approximately 730 amino acids in the average polypeptide but can vary dependent on the protein that is made) • Only certain genes in a genome need to be expressed depending on: • Cell specialism • Environment • Therefore not all genes (are transcribed) and translated • If a cell needs to produce a lot of a certain protein (e.g. β cells in the pancreas specialize in secreting insulin to control blood sugar) then many copies of the required mRNA are created.
- 109. 2.7 U.7 Codons of three bases on mRNA correspond to one amino acid in a polypeptide. The genetic code is the set of rules by which information encoded in mRNA sequences is converted into proteins (amino acid sequences) by living cells • Codons are a triplet of bases which encodes a particular amino acid • As there are four bases, there are 64 different codon combinations (4 x 4 x 4 = 64) • The codons can translate for 20 amino acids based on Amino acids are carried by transfer RNA (tRNA) The anti-codons on tRNA are complementary to the codons on mRNA • Different codons can translate for the same amino acid (e.g. GAU and GAC both translate for Aspartate) therefore the genetic code is said to be degenerate • The order of the codons determines the amino acid sequence for a protein • The coding region always starts with a START codon (AUG) therefore the first amino acid in all polypeptides is Methionine • The coding region of mRNA terminates with a STOP codon - the STOP codon does not add an amino acid – instead it causes the release of the polypeptide
- 110. 2.7 S.1 Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid. 2.7 S.3 Use a table of mRNA codons and their corresponding amino acids to deduce the sequence of amino acids coded by a short mRNA strand of known base sequence. 2.7 S.4 Deducing the DNA base sequence for the mRNA strand. The diagram summarizes the process of protein synthesis. You should be able to use a section of genetic code, transcribe and translate it to deduce the polypeptide synthesized.
- 111. 2.7.S1, 2.7.S3, 2.7.S4 Practice transcribing and translating using the learn genetics tutorial. http://learn.genetics.utah.edu/content/molecules/transcribe/
- 112. 2.7 S.1, 2.7 S.3, 2.7 S.4 Now use this table to answer the questions on the next slide n.b. You just have to be able to use the table. You do not have to memorize which codon translates to which amino acid.
- 113. 2.7 S.1, 2.7 S.3, 2.7 S.4 1. Deduce the codon(s) that translate for Aspartate. 2. If mRNA contains the base sequence CUGACUAGGUCCGGA a. deduce the amino acid sequence of the polypeptide translated. b. deduce the base sequence of the DNA antisense strand from which the mRNA was transcribed. 3. If mRNA contains the base sequence ACUAAC deduce the base sequence of the DNA sense strand.
- 114. 1. Deduce the codon(s) that translate for Aspartate. 2. If mRNA contains the base sequence CUGACUAGGUCCGGA a. deduce the amino acid sequence of the polypeptide translated. b. deduce the base sequence of the DNA antisense strand from which the mRNA was transcribed. 3. If mRNA contains the base sequence ACUAAC deduce the base sequence of the DNA sense strand. 2.7 S.1, 2.7 S.3, 2.7 S.4
- 115. 2.7 S.1, 2.7 S.3, 2.7 S.4 1. Deduce the codon(s) that translate for Aspartate. 2. If mRNA contains the base sequence CUGACUAGGUCCGGA a. deduce the amino acid sequence of the polypeptide translated. b. deduce the base sequence of the DNA antisense strand from which the mRNA was transcribed. 3. If mRNA contains the base sequence ACUAAC deduce the base sequence of the DNA sense strand. (the sense strand is the template for the mRNA the only change is that uracil is replaced by thymine) ACTAAC GACTGATCCAGGCCT (the antisense strand is complementary to the mRNA, but remember that uracil is replaced by thymine) GAU, GAC Leucine + Threonine + Lysine + Arginine + Serine + Glycine
- 116. 2.7 S.1 Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid.
- 117. 2.7 S.1, 2.7 S.3, 2.7 S.4
- 118. 2.7 S.1 Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid.
- 119. 2.7 S.1, 2.7 S.3, 2.7 S.4
- 120. 2.7 S.1 Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid.
- 121. 7.3 U.4 Free ribosomes synthesize proteins for use primarily within the cell. https://www.studyblue.com/notes/note/n/molecular-exam-3/deck/2630328 • Free ribosomes in the cytoplasm synthesize proteins that will be used inside the cell in the cytoplasm, mitochondria and chloroplasts (in autotrophs)
- 122. Ribosomes effect in translation • Ribosome are found in Prokaryotes (70's) and Eukaryotes (80's). Including P and A sites. START codons and STOP codons begin and termination translation. Polyribosome= Polysomes • Multiple ribosomes on the same mRNA at the same time. • All ribosome move 5' to 3' in sequence. • In protein synthesis polyribosomes increase the quantity of polypeptide synthesized. 7.3 S.1 Identification of polysomes in electron micrographs of prokaryotes and eukaryotes.
- 123. 7.3 U.4 Free ribosomes synthesize proteins for use primarily within the cell.
- 124. 7.3 S.1 Identification of polysomes in electron micrographs of prokaryotes and eukaryotes. • In prokaryotes, several ribosomes can attach themselves to the growing mRNA chains to form a polysome while the mRNA chains are still attached to the DNA
- 125. 7.3 S.1 Identification of polysomes in electron micrographs of prokaryotes and eukaryotes. • In eukaryotes, the mRNA detaches from the DNA and is then transported through pores in the nuclear envelope to the ribosomes in the cytoplasm. Once in the cytosol, eukaryote mRNA can also form polysomes
- 126. 7.3 U.5 Bound ribosomes synthesize proteins primarily for secretion or for use in lysosomes. • Ribosomes attached to ER create proteins that are secreted from the cell by exocytosis or are used in lysosomes. • Proteins that are destined to be used in lysosomes, ER, Golgi Apparatus, the plasma membrane or secreted by the cell are made by ribosomes bound by the endoplasmic reticulum • Ribosomes that become bound to the ER are directed here by a signal sequence that is part of that specific polypeptide • This signal sequence on the polypeptide binds to a signal recognition protein (SRP) • The SRP guides the polypeptide and ribosome to the ER where it binds to an SRP receptor http://herbmitchell.info/Figure.4-8-Synthesissecretoryprotein.jpg
- 127. 7.3 U.6 Translation can occur immediately after transcription in prokaryotes due to the absence of a nuclear membrane. • Since prokaryotic DNA is not compartmentalized into a nucleus, once transcription begins creating a strand of mRNA, translation can begin immediately as the mRNA strand is created • In eukaryotes, the completed mRNA has to be transported from the nucleus, through the nuclear pore to the ribosome on the ER or in the cytosol http://www.mun.ca/biology/scarr/iGen3_05-09_Figure-L.jpg
- 128. Prokaryotes vs. Eukaryotes Prokaryotes Eukaryotes • Transcription and translation both in cytoplasm • DNA/RNA in cytoplasm • RNA poly binds directly to promoter • Transcription makes mRNA (not processed) • No introns • Transcription in nucleus; translation in cytoplasm • DNA in nucleus, RNA travels in/out nucleus • RNA poly binds to TATA box & transcription factors • Transcription makes pre- mRNA RNA processing final mRNA • Exons, introns (cut out) 7.3 U.6 Translation can occur immediately after transcription in prokaryotes due to the absence of a nuclear membrane.
- 129. Structure of Proteins The complex structure of proteins is explained by referring to 4 levels of organization A. Primary B. Secondary C. Tertiary D. Quaternary http://upload.wikimedia.org/wikipedia/commons/2/26/225_Peptide_Bond-01.jpg
- 130. Structure of Proteins Primary structure: • The order/ number of amino acids in a polypeptide chain. • Linear shape (no internal bonding) 7.3 U.7 The sequence and number of amino acids in the polypeptide is the primary structure.
- 131. 7.3 U.8 The secondary structure is the formation of alpha helices and beta pleated sheets stabilized by hydrogen bonding Secondary Structure: Hydrogen bonding causes The primary structure of the polypeptide to fold and coil Into some characteristic ways: • Alpha Helix • Beta pleated sheets
- 132. Beta-pleated sheets: • Flat, zig-zag structure • A number of chains which are hydrogen bonded together • Forms a sheet Example: Fibers in in silk 7.3 U.8 The secondary structure is the formation of alpha helices and beta pleated sheets stabilized by hydrogen bonding
- 133. • Tertiary structure is the three-dimensional conformation of a polypeptide. • The polypeptide folds just after it is formed in translation. • The shape is maintained by intermolecular bonds 7.3 U.9 The tertiary structure is the further folding of the polypeptide stabilized by interactions between R groups. http://cnx.org/resources/36c08f3ac1c144763610fa69fbb9e278/Figure_03_04_08.jpg
- 134. 7.3 U.10 The quaternary structure exists in proteins with more than one polypeptide chain. • Quaternary structure is the linking together of two or more polypeptides to form a single protein. • The protein structure below has 4 different polypeptide chains. http://www.topsan.org/@api/deki/files/6029/=EK5976M_Fig3Comparisons.png
- 135. Conserved sequence: a homologous sequence of DNA that is identical across all members of a species. Bioinformatics: uses computer databases to store and analyze gene & protein sequences from large amounts of data collected from sequencing genes of various organisms Faster, more powerful computers allow scientist to identify conserved sequences & genes by looking for patterns and homologous sequences within organisms’ genome. If a sequence is homologous across species or individuals of a species, it usually has a functional role. Eg. It codes for a protein (a gene). 7.3 S.2 The use of molecular visualization software to analyze the structure of eukaryotic ribosomes and a tRNA molecule.
- 136. 2.5 Enzymes Essential idea: Enzymes control the metabolism of the cell. http://cdn.instructables.com/F7F/38MA/HAFHKT7I/F7F38MAHAFHKT7I.LARGE.jpg Below is an enzymatic reaction browning, which may protect the developing seeds from pathogens
- 137. 8.1 Metabolism (AHL) Essential idea: Metabolic reactions are regulated in response to the cell’s needs. https://mediaeatout.files.wordpress.com/2013/11/candidates-eating-obama-sized.jpg
- 138. 2.5 U.1 Enzymes have an active site to which specific substrates bind. 2.5 U.2 Enzyme catalysis involves molecular motion and the collision of substrates with the active site. Enzyme: A globular protein that increases the rate of a biochemical reaction by lowering the activation energy threshold (i.e. a biological catalyst). The energy need for chemical reactions to occur http://www.northland.cc.mn.us/biology/biology1111/animations/enzyme.swf Use the animation to find out more about enzymes and how they work. A good alternative is How Enzymes Work from McGraw and Hill http://highered.mheducation.com/sites/0072495855/student_view0/chapter2/animation__how_enzymes_work.html
- 139. 8.1 U.2 Enzymes lower the activation energy of the chemical reactions that they catalyze. • The substrate binds to the enzymes’ active site and the active site is altered reaching the transition state (the enzyme-substrate complex). • Due to the binding the bonds in the substrate molecule are stressed/become less stable. • Binding to an enzyme lowers the overall energy needed for a reaction to occur. • The activation energy of the reaction is then becomes reduced. How do enzymes lower the activation energy of a reaction? http://en.wikipedia.org/wiki/Image:Induced_fit_diagram.png
- 140. • Enzymes are protein catalysts that enormously speed up reactions. They often have an “-ase” ending to their name. – e.g., hexokinase, catalase, peptidase, mutase • They are not themselves changed (except for a brief period of time) and are the same before and after a reaction. • Enzymes: 1. Lower the activation energy: this is the MOST important characteristic 2. Do not add or remove energy from a reaction 3. Do not change the equilibrium for a reaction 4. Are reused over and over 2.5 U.1 Enzymes have an active site to which specific substrates bind.
- 141. Activation Energy is the energy need to get a reaction started. Enzymes decrease the barrier to starting a reaction. This energy is called the activation energy, or the energy required for a reaction to start. 8.1 U.2 Enzymes lower the activation energy of the chemical reactions that they catalyzed.
- 142. Enzymes Lower EA
- 143. Lock & Key Hypothesis a) Large globular protein enzyme b) Active Site where the substrate combines to the enzyme c)Substrate which fits the active site d) Activated complex The substrate is weakened to allow the reaction. e) Unchanged enzyme/ re-used at low concentrations f) Product of the reaction * Polar regions on the enzyme’s active site help bring the enzyme and the product together
- 144. Induced Fit Hypothesis Many enzymes catalyze more then one reaction, this is due to Hydrogen bonding (weak IMF’) holding the active site together. a. As the substrate approaches, polar regions of the enzyme’s active site attract the substrate. b. Enzyme activation site allows for a shape change or induce a fit. Causing a chemical change c. This stress reduces the activation energy or the reaction returns the enzyme to its original shape
- 145. 2.5 U.2 Enzyme catalysis involves molecular motion and the collision of substrates with the active site. http://www.kscience.co.uk/animations/model.swf The simulation from KScience allows you to both see enzyme kinetics happening and secondly how it is affected by different factors • Two substances must have the proper alignment and energy (in the form of motion) to create a chemical reaction • The direction and movement is constantly changing and is random • Collisions occur at random between the substrate and enzyme • Successful reactions only occur if the substrate and the active site of the enzyme are correctly aligned and the collide with sufficient KE
- 146. 2.5 U.2 Enzyme catalysis involves molecular motion and the collision of substrates with the active site. • Most enzyme reactions occur when the substrates are dissolved in water • All molecules dissolved in water are in random motion, with each molecule moving separately • If not immobilized the enzyme can move too, however enzymes tend be larger than the substrate(s) and therefore move more slowly
- 147. Maltase
- 148. 2.5 U.5 Immobilized enzymes are widely used in industry. http://pubs.rsc.org/services/images/RSCpubs.ePlatform.Service.FreeContent.ImageService.svc/ImageService/Articleimage/2013/CS/c3cs35506c/c3cs35506c-f1.gif Advantages of enzyme immobilization: • Concentration of substrate can be increased as the enzyme is not dissolved – this increases the rate of reaction • Recycled enzymes can be used many times, immobilized enzymes are easy to separate from the reaction mixture, resulting in a cost saving. • Separation of the products is straight forward (this also means that the the reaction can stopped at the correct time). • Stability of the enzyme to changes in temperature and pH is increased reducing the rate of degradation, again resulting in a cost saving. Enzymes used in industry are usually immobilized. They are attached to a material so that their movement is restricted. Common ways of doing this are: • Aggregations of enzymes bonded together • Attached to surfaces, e.g. glass • Entrapped in gels, e.g. alginate gel beads
- 149. 2.5 U.5 Immobilized enzymes are widely used in industry. Common uses of enzymes in industry include: http://pubs.rsc.org/services/images/RSCpubs.ePlatform.Service.FreeContent.ImageService.svc/ImageService/Articleimage/2013/CS/c3cs35506c/c3cs35506c-f1.gif 1.Detergents 2.Biofuels 3.Textiles 4.Brewing 5.Medicine & Biotechnology 6.Juice yield 7.Paper production
- 150. 1. Detergents contain proteases and lipases to help breakdown protein and fat stains https://i1.ytimg.com/vi/lQ6fCZgYc8g/hqdefault.jpg
- 151. 2. Enzymes are used to breakdown the starch in grains into biofuels that can be combusted http://chromblog.thermoscientific.com/Portals/49739/images/biofuel9.jpg http://greenodin.com/wp-content/uploads/2014/12/GreenOdin-GO-Biodiesel-Van-1200.png
- 152. 3. In the textiles industry enzymes help in the processing of fibers, e.g. polishing cloth to make it appear more shiny
- 153. 4. In the brewing industry enzymes help a number of processes including the clarification of the beerhttp://www.brewreviewcrew.com/cans-vs-bottles-fight/
- 154. 5. In Medicine & Biotechnology enzymes are widely used in everything from diagnostic tests tests to contact lens cleaners to cutting DNA in genetic engineering. http://www.medwesteye.com/wp-content/uploads/2014/12/learn-the-proper-care-of-contact-lenses.jpg
- 155. 6. Enzymes are widely used in the food industry, e.g. • fruit juice, pectin to increase the juice yield from fruit • Fructose is used as a sweetener, it is converted from glucose by isomerase • Rennin is used to help in cheese productionhttps://theramblingreed.files.wordpress.com/2014/08/img_6338.jpg
- 156. 7. Paper production uses enzymes to helping in the pulping of wood http://i00.i.aliimg.com/img/pb/479/389/262/1281752366918_hz-cnmyalibaba-web2_15708.jpg
- 157. Optimal environmental condition(s) favor the most active enzyme conformation. • Temperature • pH • Concentration of substrate/enzyme • Cofactors • Enzyme inhibitors • Allosteric regulation (noncompetitive) • Ionic concentration 2.5 U.3 Temperature, pH and substrate concentration affect the rate of activity of enzymes.
- 158. 1. The Effect Of Temperature (a) •Increase Kinetic energy of substrate and enzyme. •Increased chance of collisions •Low temperatures has a low rate of reaction (b) •Optimum temperature = maximum rate of reaction •Balance between enzyme stability and kinetic energy of reactants (c) •Rapid decrease in the rate of reaction •High temperatures destabilize the enzyme molecule •Enzyme is denatured
- 159. Denaturing is a structural change in a protein that results in a loss (usually permanent) of its biological properties. • Enzymes are globular proteins • Enzymes have tertiary structure • Tertiary structure is maintained by hydrogen, ionic and covalent bonds • Shape of the active site is maintained by hydrogen, ionic and covalent bonds The bonds within enzymes (and proteins) has an increasing strength of: Hydrogen Ionic Covalent 2.5 U.4 Enzymes can be denatured.
- 160. Proteins found in the egg white include albumins, globulins and mucoproteins Under intense heat, hydrogen bonds that formed during the secondary structure are broken The proteins then lose their shapes, thus changing their functions By cooking it, you have effectively denatured the egg What happens when you cook an egg?
- 161. 2. The Effect Of pH (a) Decrease in pH (increase in H+) • H+ interact with exposed R groups on active site. • Enzyme active site changes shape • Specificity reduced • Decrease in rate of reaction (b) Optimum rate of reaction for the pH= (d) • Active site structure and structure specific to the complementary shape of the substrate. • Successful activated complex and therefore reactions occur. (c) Increase in pH (increase in OH-) • Increase in base (OH-) concentration • Enzyme active site changes shape • Specificity reduced • Decrease in rate of reaction
- 162. Denaturation of an enzyme by a change in pH • If there is a deviation from the optimal pH the hydrogen sulfide bridges break and the enzyme loses shape. • Loss of the activation site shape leads to loss of function
- 163. Denaturation of an enzyme by a change in pH • Enzymes have an optimum pH at which they achieve their maximum rate of reaction • Pepsin has an optimal pH of 2 at (a) • Amylase has an optimal pH of (c) • pH affects the charge of the amino acids of the active site • This changes the properties of the active site • e.g. carboxyl R group will be uncharged COOH at low pH but COO- at high pH.
- 164. 3. The effect of substrate concentration (a) •Increase conc. of substrate molecules •Increased chance of collision with enzyme •Greater chance of forming activated complex •Increase in rate of reaction (b) •Rate begins to level •Active sites beginning to become saturated with substrate (fully occupied) •New substrate must wait for previous reaction to complete and the product to exit the active site (c) •Full saturation of the active sites by substrate •Rate becomes constant for further increases in substrate concentration.
- 165. • Metabolism: the sum total of all chemical reactions that occur within an organism. • Metabolic Pathway sequences of biochemical reactions that occur in all living cells Two types of metabolic pathways 1. Linear Metabolic Pathways: •Chemical changes in living things often occurring with a number of intermediate stages. •Each stage has its own enzyme. •Catabolic pathways breakdown molecules •Anabolic pathways build up molecules 8.1 U.1 Metabolic pathways consist of chains and cycles of enzyme- catalyzed reactions. The first step of cellular respiration Glycolysis
- 166. 2. Cyclic Metabolic Pathways: •The initial substrate is fed into the cycle. • Enzyme (a) combines the regenerated Intermediate 4 to catalyzes the production of intermediate 1 • Enzyme (b) converts intermediate 1 to intermediate 2 • Enzyme (c) converts intermediate 2 to intermediate 3. The product is formed and removed. • Enzyme (d) converts intermediate 3 to intermediate 4 and the cycle repeats.
- 167. Second step of cellular respiration Krebs Cycle
- 168. • Inhibitors are substances that reduce or completely stop the action of an enzyme • Inhibition can act on the active site (competitive) or on another region of the enzyme molecule(non-competitive). The competition in the former being for the active site of the enzyme. Competitive blocks the active site and non-competitive attaches to another site on the enzyme and changes the shape of the active site of the enzyme 8.1 U.3 Enzyme inhibitors can be competitive or non-competitive.
- 169. Non-Competitive inhibition Competitive Inhibition 8.1 U.3 Enzyme inhibitors can be competitive or non-competitive.
- 170. 8.1 S.2 Distinguishing different types of inhibition from graphs at specified substrate concentration. https://wikispaces.psu.edu/download/attachments/46924781/image-6.jpg Rate of reaction is reduced Features of competitive inhibitors When the concentration of substrate begins to exceed the amount of inhibitor, the maximum rate of the uninhibited enzyme can be achieved. However, it takes a much higher concentration of substrate to achieve this maximum rate.
- 171. https://wikispaces.psu.edu/download/attachments/46924781/image-6.jpg Rate of reaction is reduced Features of non-competitive inhibitors It takes approximately the same concentration of enzyme to reach the maximum rate, but the maximum rate is lower than the uninhibited enzyme. • The binding of the non-competitive inhibitor prevents some of the enzymes from being able to react regardless of substrate concentration. • Those enzymes that do not bind inhibitors follow the same pattern as the normal enzyme. 8.1 S.2 Distinguishing different types of inhibition from graphs at specified substrate concentration.
- 172. 8.1 A.1 End-product inhibition of the pathway that converts threonine to isoleucine. http://www.uic.edu/classes/bios/bios100/lecturesf04am/feedback-inh.gif Isoleucine from threonine • The synthesis of isoleucine from threonine in a series of five enzyme- catalysed steps • As the concentration of isoleucine increases, some of it binds to the allosteric site the enzyme threonine deaminase acts as a non-competitive inhibitor. • The pathway is then turned off, regulating isoleucine production.
- 173. Isoleucine in the body is responsible for some of the following: energy levels, sugar levels, hemoglobin production. The amino acid has been know to assist in wound healing, simulating immune function and promoting the secretion of several important hormones. 8.1 A.1 End-product inhibition of the pathway that converts threonine to isoleucine.
- 174. 8.1 A.2 Use of databases to identify potential new anti-malarial drugs. http://upload.wikimedia.org/wikipedia/commons/0/02/Mosquito_bite4.jpg • Malaria is a disease caused by the pathogen Plasmodium falciparum. • This protozoan uses mosquitoes as a host as well as humans and hence can be passed on by mosquito bites
- 175. • In one study, approx. 300,000 chemicals were screened against a chloroquine-sensitive 3D7 strain and the chloroquine-resistant K1 strain of P. falciparum. • Other related and unrelated organisms, including human cell lines, were also screened. • (19) new chemicals that inhibit the enzymes normally targeted by anti-malarial drugs were identified • Additionally (15) chemicals that bind to malarial proteins were identified – this can help in the location of P. falciparum • These results indicate possible new directions for drug research. Increasing drug resistance to anti-malarial drugs has lead to the use of bioinformatics and chemogenomics to try and identify new drugs. 8.1 A.2 Use of databases to identify potential new anti-malarial drugs.
- 176. • Sometimes when a chemical binds to a target site, it can significantly alter metabolic activity. • Massive libraries of chemicals are tested individually on a range of related organisms. • For each organism a range of target sites are identified. • A range of chemicals which are known to work on those sites are tested. Bioinformatics is an approach whereby multiple research groups can add information to a database enabling other groups to query the database. Bioinformatics has facilitated research into metabolic pathways is referred to as chemogenomics.
- 177. 8.1 S.1 Calculating and plotting rates of reaction from raw experimental results. The rate of reaction can be calculated using the formula: Rate of reaction (s-1) = 1 / time taken (s) Time taken in enzyme experiments this is commonly the time to reach a measurable end point or when a standard event, caused by the enzyme reaction, has come to pass. This is usually measured by the effects of the accumulation of product, but can as easily be measured by the disappearance of substrates. http://www.scienceexperimentsforkids.us/wp-content/uploads/2011/08/hydrogen-experiments-for-kids-3-img.jpg Use the results from it or data from one of your enzyme inhibition labs to calculate the rate of reaction. Enzyme inhibition can be investigated using these two outlines by Science & Plants for Schools: • The effect of end product, phosphate, upon the enzyme phosphatase • The inhibition of catechol oxidase by lead
- 178. Lactose Intolerance • Lactose (milk sugar) can cause allergies in some people. • This is often because they are unable to produce the enzyme lactase in sufficient quantities. • Most people produce less lactase as they get older. After all, we don’t live off milk once we have been weaned. • In some regions such as Europe, a mutation has allowed lactose production to continue into adulthood. This mutation is not present in people who are lactose intolerant 2.5 A.1 Methods of production of lactose-free milk and its advantages.
- 179. Green High Tolerance Red Low Tolerance Global estimates of lactose intolerance.
- 180. How can we cope with lactose intolerance? • Take a lactase supplement. These are produced industrially using the Aspergillus niger fungus • Drink lactose free milk. Milk treated with lactase (produced by A. niger) and essentially ‘pre-digested’ before being packaged.
- 181. 2.5 A.1 Methods of production of lactose-free milk and its advantages. Other uses of lactose free milk: • As a means to increase the sweetness of milk (glucose and galactose are sweeter in flavor), thus negating the need for artificial sweeteners • As a way of reducing the crystallization of ice-creams (glucose and galactose are more soluble than lactose) • As a means of shortening the production time for yogurts or cheese (bacteria ferment glucose and galactose more readily than lactose) Production of Lactose-free milk • Lactase obtained from commonly from yeast (bacteria is an alternative) • Lactase is bound to the surface of alginate beads • Milk is passed (repeatedly) over the beads • The lactose is broken down into glucose and galactose • The immobilized enzyme remains to be used again and does not affect the quality of the lactose free milk
- 182. 2.5 S.1 Design of experiments to test the effect of temperature, pH and substrate concentration on the activity of enzymes. 2.5 S.2 Experimental investigation of a factor affecting enzyme activity. (Practical 3) Possible research questions, what are you going to investigate (independent variable)? • What is the effect of substrate concentration? • What is the effect of temperature? • What is the effect of pH? • Which type of yeast has a higher concentration of catalase? Important things to consider: • How are you going to vary the mass/volume/concentration of your variable? • What units will you be measuring your variable in? • Have you chosen an effect range or values to answer your question? • Are the concentrations/chemicals you are using safe to handle? Catalase is one of the most widespread enzymes. It catalyzes the conversion of hydrogen peroxide, a toxic by-product of metabolism, into water and oxygen.
- 183. How are you going to measure your results (dependent variable)? • Are you measuring the increase of a product or the disappearance of a substrate? • Are you measuring directly (e.g. testing for the concentration of the product) or indirectly (change in pH)? • What equipment will you be using to measure your results? • What are the units and uncertainty given both the equipment and how you choose to use it? • What time period do you need to run the experiment for? How fast is the enzyme action likely to be? • How many repeats will you need to make sure your results are reliable? 2.5 S.1 Design of experiments to test the effect of temperature, pH and substrate concentration on the activity of enzymes. 2.5 S.2 Experimental investigation of a factor affecting enzyme activity. (Practical 3)
- 184. How are you going to make sure it is a fair test (control variables)? • What variables other than your independent variable could affect the results? • Why would these variables affect the results? • How will you ensure each is kept constant and monitored? • What level should they be kept constant at? If a control variable is too far from it’s optimum then it could limit the enzyme action and no change would be seen in the results. • If a variable cannot be controlled it should still be discussed and considered as an uncontrolled variable. Safety and ethics: • Are you using any equipment that may cause you or others harm? What steps have you taken to minimize this risk? • If you intend to use animals have you first considered alternative subjects? • If you still intend to use animals are subjects have you ensured both: o no harm comes to them as a result of the experiment o The experiment does not induce stress or conditions beyond that normally found in their natural environment 2.5 S.1 Design of experiments to test the effect of temperature, pH and substrate concentration on the activity of enzymes. 2.5 S.2 Experimental investigation of a factor affecting enzyme activity. (Practical 3)
